- EN - English
- CN - 中文
Live-cell Imaging by Super-resolution Confocal Live Imaging Microscopy (SCLIM): Simultaneous Three-color and Four-dimensional Live Cell Imaging with High Space and Time Resolution
超分辨率共聚焦显微镜活体细胞成像(SCLIM):高时空分辨率的三色、四维同步活体细胞成像
发布: 2020年09月05日第10卷第17期 DOI: 10.21769/BioProtoc.3732 浏览次数: 5978
评审: Chiara AmbrogioSneha Ramesh ManiAnonymous reviewer(s)
Abstract
Many questions in cell biology can be solved by state-of-the-art technology of live cell imaging. One good example is the mechanism of membrane traffic, in which small membrane carriers are rapidly moving around in the cytoplasm to deliver cargo proteins between organelles. For directly visualizing the events in membrane trafficking system, researchers have long awaited the technology that enables simultaneous multi-color and four-dimensional observation at high space and time resolution. Super-resolution microscopy methods, for example STED, PALM/STORM, and SIM, provide greater spatial resolution, however, these methods are not enough in temporal resolution. The super-resolution confocal live imaging microscopy (SCLIM) that we developed has now achieved the performance required. By using SCLIM, we have conducted high spatiotemporal visualization of secretory cargo together with early and late Golgi resident proteins tagged with three different fluorescence proteins. We have demonstrated that secretory cargo is indeed delivered within the Golgi by cisternal maturation. In addition, we have visualized details of secretory cargo trafficking in the Golgi, including formation of zones within a maturing cisterna, in which Golgi resident proteins are segregated, and movement of cargo between these zones. This protocol can be used for simultaneous three-color and four-dimensional observation of various phenomena in living cells, from yeast to higher plants and animals, at high spatiotemporal resolution.
Keywords: Live cell imaging (活体细胞成像)Background
Development of live cell imaging by state-of-the-art light microscopy has greatly contributed to address many problems remaining in cell biology. For example, to visualize the events in secretory pathway, in which small membrane carriers are rapidly moving around in the cytoplasm, requires high speed and three-dimensional (3D) image acquisition. Super-resolution confocal live imaging microscopy (SCLIM), which we have developed (Kurokawa et al., 2013), can achieve high speed visualization of dynamic behavior of 3D structures tagged with green and red fluorescence proteins in living cells with spatial resolution beyond the diffraction limit of light (Matsuura-Tokita et al., 2006; Ito et al., 2012; Okamoto et al., 2012; Suda et al., 2013; Uemura et al., 2014; Kurokawa et al., 2014; Iwai et al., 2016; Ishii et al., 2016 and 2019; Kurokawa et al., 2016). We can now use 3-color version of SCLIM, which enable us to conduct simultaneous 3-color (green, red, and infrared) observation in living yeast, Drosophila, plant, and mammalian cells at high spatial and time resolution (Ito et al., 2018; Kurokawa et al., 2019; Maeda et al., 2019; Tojima et al., 2019; Fujii et al., 2020). We have been working on the molecular mechanisms underlying protein transport and sorting in the Golgi apparatus and have utilized the cutting-edge performance of our technology to tackle a big question in this field. The purpose of this article is to introduce to readers our approaches by super-resolution and high-speed live cell imaging and provide basic protocols.
The Golgi apparatus is the central station of membrane traffic. One third of proteins in eukaryotic cells are newly synthesized in the endoplasmic reticulum (ER) and then delivered to the Golgi. In the Golgi, they are processed and glycosylated before being sorted to their final destinations (Emr et al., 2009). The Golgi is usually in the form of ordered stacks of several cisternae. Secretory cargo travels across the stack from the cis side to the trans side of cisternae and then to the trans-Golgi network (TGN) (Griffiths and Simons, 1986). The budding yeast Saccharomyces cerevisiae presents a unique example of the Golgi, in which individual cisternae, cis, medial and trans, do not stack but scatter in the cytoplasm (Matsuura-Tokita et al., 2006; Okamoto et al., 2012). Three major models of secretory cargo traffic within the Golgi have been proposed: 1) traffic via cisternal maturation, 2) traffic by anterograde vesicular carriers, and 3) traffic via interconnected continuity of cisternae (Glick and Nakano, 2009; Nakano and Luini, 2010; Pfeffer, 2010; Glick and Luini, 2011). Among these models, the cisternal maturation model has been favored to explain the live imaging observation in budding yeast that Golgi cisternae mature from early to late cisternae (Losev et al., 2006; Matsuura-Tokita et al., 2006; Rivera-Molina and Novick, 2009) and the movement of large cargo through the Golgi stacks without leaving cisternae in mammals (Bonfanti et al., 1998; Lanoix et al., 2001; Martinez-Menarguez et al., 2001; Mironov et al., 2001).The lacking proof of this mechanism was to visualize the delivery of cargo remaining in the maturing Golgi cisterna.
Electron microscopy shows detailed snap information where secretory cargo and Golgi resident proteins localize in the individual Golgi cisternae, but cannot directly visualize how secretory cargo traverse between these cisternae. Live imaging by using conventional epi- and confocal fluorescence microscopies visualize behavior of secretory cargo in vivo, but cannot provide detailed information where secretory cargo and Golgi resident proteins locate within the individual Golgi cisternae because of low spatial resolution. Super-resolution microscopy methods, for example STED, PALM/STORM, and SIM overcoming the diffraction limit of light, provide greater spatial resolution (Schermelleh et al., 2010). However, these methods are not high in temporal resolution, because time and space resolutions usually trade off each other. By using SCLIM technology, we have shown that secretory cargo is delivered within the Golgi by cisternal maturation. Maturing cisterna involves segregated zones of the earlier and later Golgi resident proteins. The location of cargo changes from the early to the late zone within the cisterna during the progression of maturation (Kurokawa et al., 2019).
Materials and Reagents
- 90 mm diameter Cell culture dishes, sterile (Iwaki, catalog number: SH90-15 )
- 22 mm x 40 mm Micro cover glasses (Matusnami Glass, thickness No.1, 0.13-0.17mm)
- 18 mm x 18 mm Micro cover glasses (Matusnami Glass, thickness No.1, 0.13-0.17mm)
- 20 ml Syringes (Terumo, catalog number: SS-20ESZ )
- Pipette tips, sterile (Sorenson, 1-200 µl Graduated tip, catalog number: 27730 )
- Kimwipes (Nihon Seishi, catalog number: 62011 )
- Tissue culture test plates, 24-well plate, sterile (TPP, catalog number: 92424 )
- Toothpicks sterilized before use (any brand)
- Budding yeast cells expressing two Golgi markers and secretory cargo tagged with different fluorescent proteins. (see Procedure B for details)
- Silicon grease (Dow Corning Toray, catalog number: HVG-50 TUBE )
- Concanavalin A (Sigma-Aldrich, catalog number: C2010 )
- Yeast nitrogen base without amino acids (Difco Laboratories)
- Casamino acids (Difco Laboratories)
- Glucose (Nacalai Tesque)
- Agar (Wako)
- MCD agar plates (see Recipes)
- MCD medium (see Recipes)
Equipment
- Micropipette (Gilson, Pipetman, model: P200 )
- Bio Shaker (Taitec, model: M-BR-022UP )
- SCLIM (Figure 1) is equipped with:
- Inverted fluorescence microscope (Olympus, model: IX-73 )
- Objective lens (Olympus, UPlanSApo 100x NA 1.4 oil objective lens)
- Spinning-disk confocal scanner (Yokogawa Electric, CSU10 custom made model)
Other type of CSU can be used (e.g., CSU10, CSUX1, CSUW1). Confocal scanner is equipped with custom made dichroic mirror having high spectroscopic properties for 3-color observation (Kurokawa et al., 2013). - Excitation lasers: Solid-state 3 lasers with emission at 473 nm (Cobolt, CW 473nm, DPSS, 50 mW), 561 nm (Cobolt, CW 561 nm, DPSS, 50 mW), and 671 nm (CrystaLaser, CW 671 nm, DPSS, 100 mW)
These 3 lasers can excite GFP, mRFP or mCherry, and iRFP simultaneously. Irradiation of the sample by these 3 lasers is controlled by 3 independent electromagnetic shutters (Sigma Koki). These shutters are operated by the demand of the custom-made software. Irradiation powers of 3 lasers can be adjusted with variable ND filters. - Spectroscopic unit: A custom made system equipped with custom-made dichroic mirrors, reflection mirrors, band pass filters, and long pass filter to separate 3-color fluorescence (green, red, and infra-red) images to 3 different channels (Kurokawa et al., 2013)
The angles of dichroic mirrors and reflection mirrors can be adjusted from outside of the spectroscopic unit. - Magnification lens (NIKON, VM lens C-4x [4x] or C-4x plus C-2.5x [10x])
A magnification lens is putted in the light path between the confocal scanner and spectroscopic unit for raising spatial resolution. - EM-CCD cameras (Hamamatsu Photonics, C9100-13)
They contain 512 x 512 frame transfer CCD sensor. Pixel size of sensor is 16 x 16 µm. 3 EM-CCD cameras are set up for taking green, red, and infra-red fluorescence images. - Image intensifier (Hamamatsu Photonics)
3 Image intensifiers are equipped with each 3 EMCCD cameras. Image intensifiers are cooled with a custom-made cooling system to achieve low signal-to-noise ratio amplification. The amplification gain of these 3 image intensifiers are controlled independently. - Piezo actuator (Yokogawa Electric, custom made model)
It is equipped at the neck of objective lens to oscillate Z-axis position of the lens at high frequency (maximum, 30 Hz). Normally, we use it at 4-15 Hz. - Thermo-control stage (Tokai Hit)
The temperature of the sample during observation can be controlled by thermo-control stage. - Image acquisition system
Custom made system controls simultaneous collection of 3 EM-CCD camera’s images at each Z-axis position and each time point (Yokogawa Electric).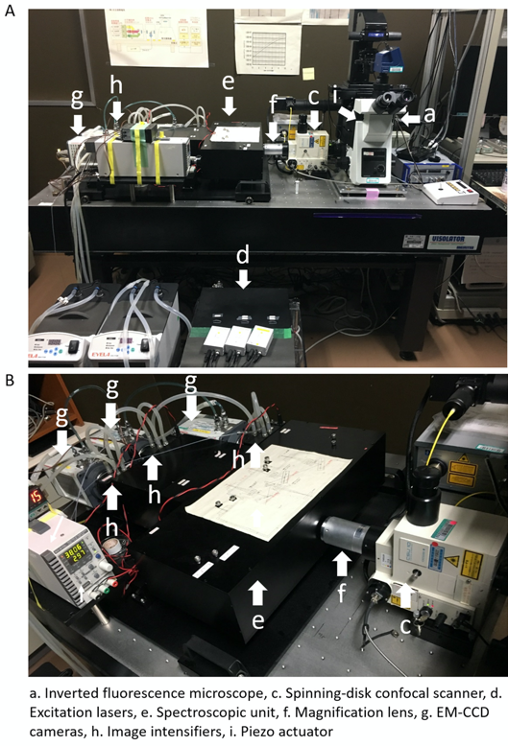
Figure 1. Set up of 3-color SCLIM. A. An inverted microscopy (a) is combined with, an ultra-fast piezo actuator (i), a custom-made spinning-disk confocal scanner (c), 3 excitation lasers (d), a magnifier lens (f), a custom-made spectroscopic unit (containing dichroic mirrors, refraction mirrors, and band-pass and long-pass filters) (e), 3 cooled image intensifiers (h), and 3 EM-CCD cameras (g). B. Another view of the custom-made spectroscopic unit (e), 3 cooled image intensifiers (h), and 3 EM-CCD cameras (g) is shown.
- Inverted fluorescence microscope (Olympus, model: IX-73 )
Software
- Volocity software (PerkinElmer)
- MetaMorph software (Molecular Devices)
Procedure
文章信息
版权信息
© 2020 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Kurokawa, K. and Nakano, A. (2020). Live-cell Imaging by Super-resolution Confocal Live Imaging Microscopy (SCLIM): Simultaneous Three-color and Four-dimensional Live Cell Imaging with High Space and Time Resolution. Bio-protocol 10(17): e3732. DOI: 10.21769/BioProtoc.3732.
- Kurokawa, K., Osakada, H., Kojidani, T., Waga, M., Suda, Y., Asakawa, H., Haraguchi, T. and Nakano, A. (2019). Visualization of secretory cargo transport within the Golgi apparatus. J Cell Biol 218(5): 1602-1618.
分类
微生物学 > 微生物细胞生物学
细胞生物学 > 细胞成像 > 活细胞成像
细胞生物学 > 细胞成像 > 超分辨率成像
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link