- EN - English
- CN - 中文
Assessing Gαq/15-signaling with IP-One: Single Plate Transfection and Assay Protocol for Cell-Based High-Throughput Assay
使用IP-One评估Gαq/ 15信号:基于细胞高通量测定的单板转染和测定方案
(*contributed equally to this work) 发布: 2020年08月20日第10卷第16期 DOI: 10.21769/BioProtoc.3715 浏览次数: 5511
评审: Marie of BoutetYasuyuki NakamuraAnonymous reviewer(s)

相关实验方案
用于比较人冷冻保存 PBMC 与全血中 JAK/STAT 信号通路的双磷酸化 CyTOF 流程
Ilyssa E. Ramos [...] James M. Cherry
2025年11月20日 2329 阅读
Abstract
Cell-based functional assays are an important part of compound screening and drug lead optimization, and they can also play a crucial role in the determination of the residues involved in ligand binding and signaling for a particular G-protein-coupled receptor. Conventional methods used for Gαq/15-coupled receptors rely on the use of fluorescent probes for Ca++ sensing (such as Fura-2 and Fluo-4) or on the incorporation of [3H]-inositol into inositol 1,4,5- triphosphate (IP3). However, these methods are not suitable for screening large libraries of compounds or for screening several mutants of the same receptor. In contrast, the IP-One assay by Cisbio is a TR-FRET assay suitable for large compound library screening when using stable cell lines that express a specific 7TMR. However, when using transiently transfected mutants of a 7TMR, this assay is not ideal, as it requires a two-step protocol of cell culture. Therefore, we have optimized the IP-One assay protocol using the reverse transfection method in 384-well plates. This offers a time- and resource-efficient alternative to the two-step protocol previously used for the screening of several mutants of Gαq/15-coupled 7TMRs.
Keywords: G-protein-coupled receptor (G蛋白偶联受体)Background
Seven-transmembrane receptors (7TMR), also known as G-protein-coupled receptors, are the most important transmembrane protein superfamily involved in signal transduction. They are the target of 30 to 50% of clinically approved drugs (Overington et al., 2006). Gαq/15-coupled 7TMRs activate phospholipase C β (PLCβ) and produce D-myo-inositol 1,4,5-triphosphate (IP3) and diacylglycerol (DAG). IP3 trigger a release of intracellular Ca++ storages and is later degraded into IP2, IP1 and ultimately D-myo-inositol (Figure 1).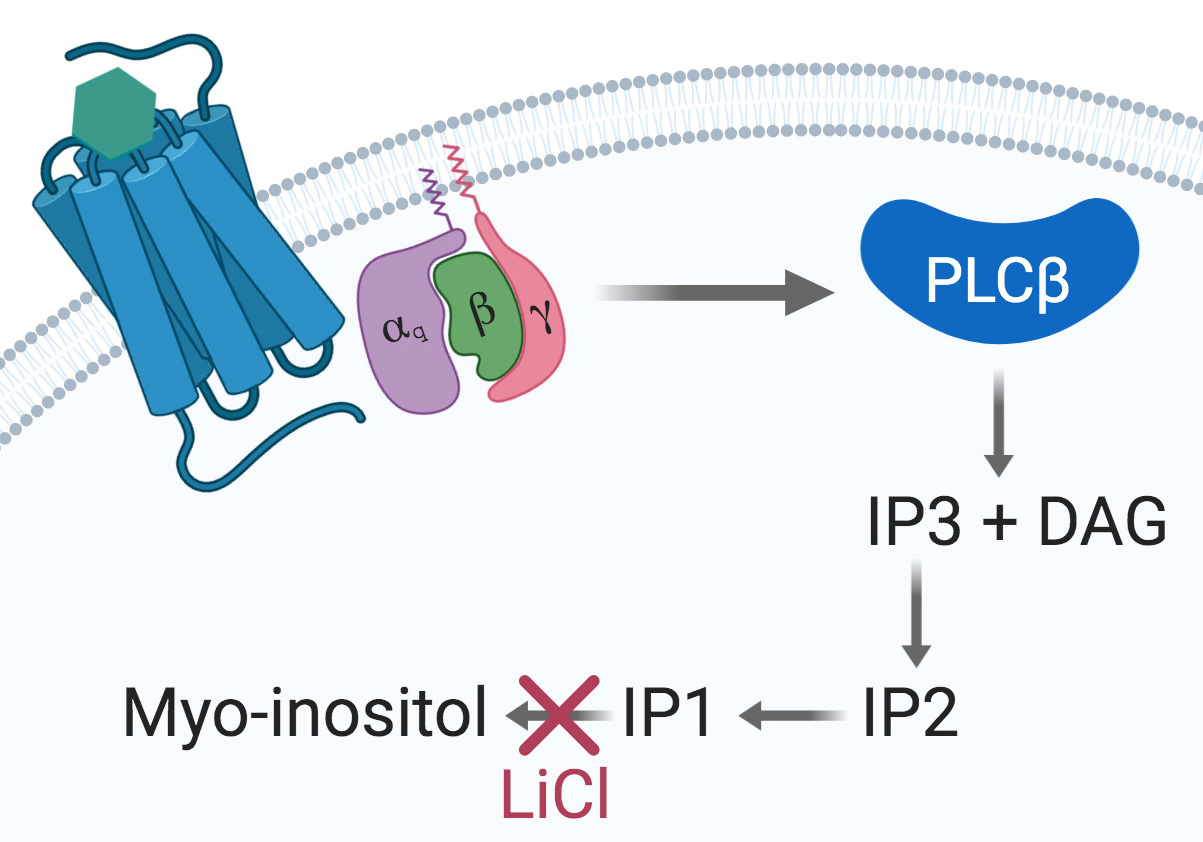
Figure 1. The Gαq signaling pathway. Upon agonist binding at a given Gαq-coupled 7TMR, the α subunit dissociates from the βγ dimer and activates the PLCβ, thus initiating the cleavage of PIP2 into IP3 and DAG. IP3 is then degraded into IP2, IP1, and myo-inositol. Accumulation of IP1 is achieved by using a stimulation buffer containing LiCl to inhibit IP1 degradation into myo-inositol and further recycling.
Gαq/15 activation is commonly measured by dectecting Ca++ flux using fluorescent calcium dye indicators. However, these indicators require either the use of an Atto-Fluor microscope with very low throughput, or investment in an expensive fluorescent imaging plate reader (FLIPR, Arkin et al., 2004). Another Gαq/15 activity quantification method relies on the detection of [3H]-inositol-phosphates ([3H]-IPs) following incubation of cells with [3H]-inositol. This highly reliable assay is based on the accumulation of [3H]-IPs after stimulation with a 7TMR agonist (Thomsen and Behan, 2007). However, both the FLIPR calcium flux assay and the [3H]-IPs detection method have important drawbacks. On the one hand, the Ca++ flux is transient and rapid, the assay does not permit the detection of constitutive activity (and thus, inverse agonism), and the readout can be interfered with by fluorescent compounds (Garbison et al., 2004). On the other hand, [3H]-IPs detection assays are time-consuming, very expensive, and generate radioactive waste that needs to be disposed of properly. A non-radioactive assay based on the detection of accumulated IP1 using a homogenous time-resolved fluorescence assay has been developed by Cisbio (Trinquet et al., 2011). This assay is based on a competition between endogenous IP1 and d2-labeled IP1 binding to an Eu cryptate-conjugated IP1 antibody (Figure 2, Liu et al., 2008).
Several protocols have been developed for the quantification of IP1 using Cisbio’s IP-One assay, but they only apply for the screening of different compounds at a single receptor. Traditional protocols for IP-One assay using transient or stable receptor expression are designed in a 2-step protocol. First, cells are plated into petri-dishes and transfected for 24 to 48 h to achieve a suitable cell-surface receptor expression (if transient expression is preferred). Next, cells are detached using trypsin and re-suspended in stimulation buffer before being distributed and stimulated into the assay 384-well plate (Shehata et al., 2016; St-Pierre et al., 2018). Other variations of this protocol using stable cell lines are reported and involve a plating step into the assay plate 24 h before stimulation (Cassutt et al., 2007; Zhang et al., 2010).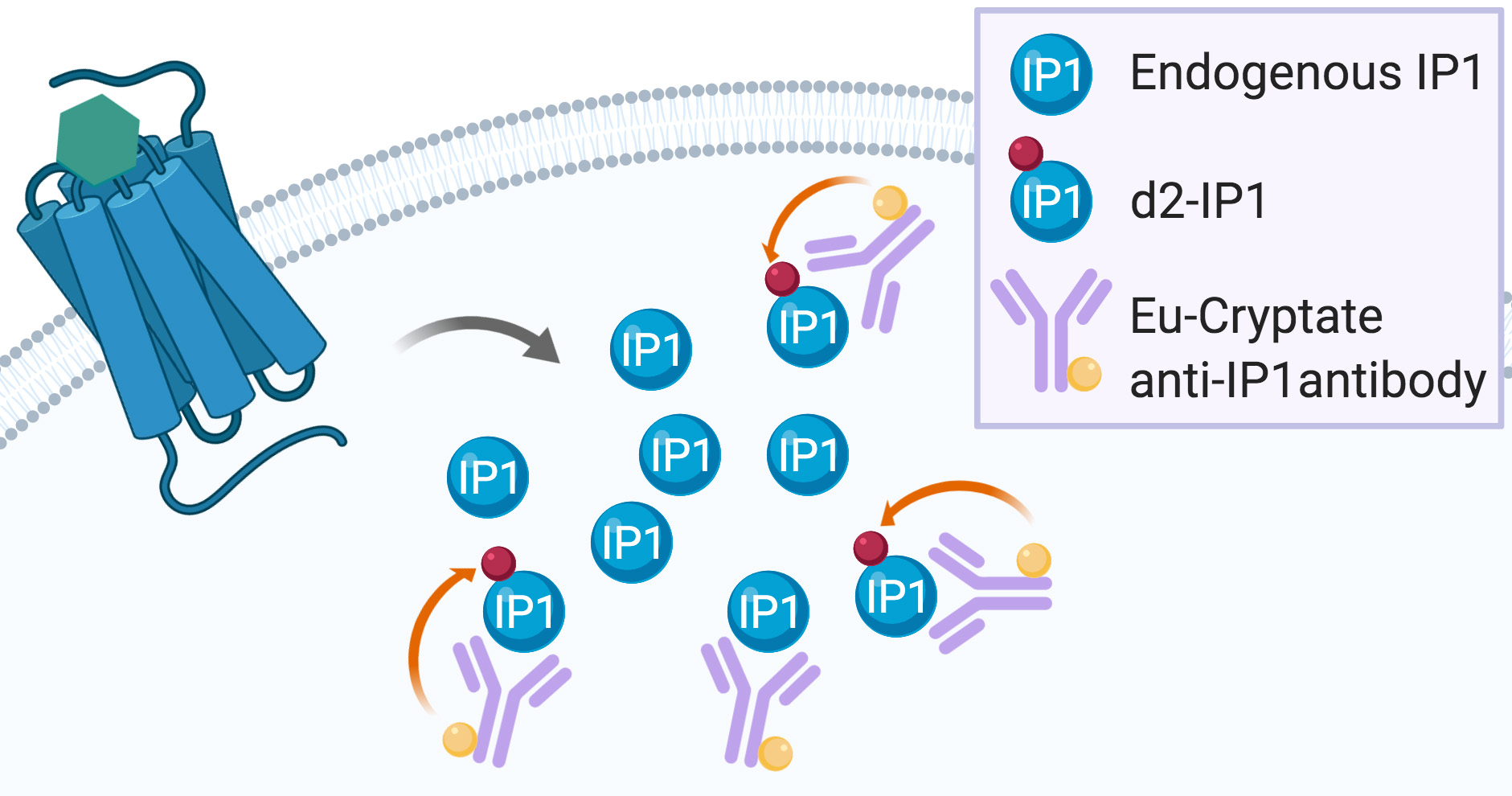
Figure 2. IP-One assay principle. Endogenous IP1 produced after 7TMR activation enter into competition with the exogenous d2-labeled IP1 for binding to the exogenous Eu-cryptate anti-IP1 antibody. When d2-IP1 is bound to the Eu-cryptate anti-IP1 antibody, the close proximity of the two fluorescent dyes allows for energy transfer during the readout (represented by the orange arrow). The more IP1 is produced by 7TMR activation, the less d2-IP1 is bound to the Eu-cryptate anti-IP1 antibody and the less energy transfer can be recorded during readout.
In order to avoid the transfection of receptors and mutants into petri dishes and then the transfer of cells into the assay plates, and to avoid unnecessary exposition to trypsin that can lead to non-functional surface receptors, we have developed a single step transfection and assay protocol. This protocol uses reverse transfection of the receptor’s DNA and direct plating of cells in a cell culture-treated and assay-suitable 384-well plate. After 48-h incubation, adherent cells are then washed and used in the IP-One assay (Gusach et al., 2019; Luginina et al., 2019). This protocol can also be used for receptors that are not coupled to Gαq by co-transfection of the Gα15/16 G protein-coding plasmid along with the receptor’s plasmid as Gα15/16 are promiscuous G proteins able to couple to non-Gαq-coupled 7TMRs (New and Wong, 2004; Robas and Fidock, 2005).
Materials and Reagents
- 10-cm cell culture-treated petri dishes (Corning, Falcon, catalog number: 353003 )
- Cell culture treated white opaque 384-well plate small volume (Greiner Bio One, catalog number: 784080 )
- Non-treated 96-well V-bottom plate 360 µl (BrandTech Scientific, catalog number: 781601 )
- Sterile 10- and 25-ml serological pipettes (Fisherbrand, catalog numbers: 14955234 [10 ml], 14955235 [25 ml])
- Sterile 1.5-ml microcentrifuge (Fisherbrand, Basix, catalog number: 509GRDSERV )
- Sterile 15-ml conical centrifuge tubes (Corning, Falcon, catalog number: C352096 )
- Sterile 50-ml conical centrifuge tubes (Corning, Falcon, catalog number: C352070 )
- Sterile 25-ml pipet basin (Fisherbrand, catalog number: 13681509 )
- Sterile 60-ml syringe with luer lock (Fisherbrand, catalog number: 14955461 )
- Sterile Uniflo Syringe Filters 0.22 µm PVDF (GE Healthcare, Whatman, catalog number: 99132502 )
- HEK293 cells (ATCC, catalog number: CRL-1573 )
- DMEM (Wisent Bioproducts, catalog number: 319-005-CL )
- Fetal bovine serum (FBS, Wisent Bioproducts, catalog number: 080-150 )
- 1 M HEPES solution (Wisent Bioproducts, catalog number: 330-050-EL )
- 200 mM L-Glutamine solution (Wisent Bioproducts, catalog number: 609-065-EL )
- 100x Penicilin-Streptomicyn solution (Wisent Bioproducts, catalog number: 450-200-EL )
- 1x PBS (Wisent Bioproducts, catalog number: 311-010-CL )
- Trypsin 0.25%-EDTA 2.21 mM (Wisent Bioproducts, catalog number: 325-043-CL )
- Opti-MEM (Gibco, catalog number: 31985062 )
- Lipofectamine 3000 (Invitrogen, catalog number: L3000001 )
- P3000 reagent (Invitrogen, includeed with Lipofectamine 3000)
- Poly-L-Lysine Hydrobromide, 25 mg (Sigma-Aldrich, catalog number: P1274-25MG )
- Moxi Z cell count cassettes Type M (Orflo, catalog number: MXC001 )
- IP-One–Gq kit 20,000 points (PerkinElmer, Cisbio, catalog number: 62IPAPEC )
- Poly-L-Lysine 0.1 mg/ml solution (see Recipes)
- Complete DMEM (media for HEK293 cells) (see Recipes)
Equipment
- Moxi Z mini automated cell counter (Orflo, catalog number: MXZ001 )
- 8-channel 10-100 µl multipipette (Eppendorf, Research plus, catalog number: 3125000036 )
- Single channel repeater pipette (Eppendorf, Repeater M4, catalog number: 4982000322 )
- HTRF filter set (λex. 320 nm, λem.1 620 nm, λem.2 665 nm)
- HTRF compatible multi-mode plate reader (Tecan, GENios Pro)
Software
- XFluor4 Excel Macro (Tecan Application to control the GENios Pro plate reader, Tecan)
- Excel 2016 (Microsoft Office, https://products.office.com)
- Prism8 (GraphPad, https://www.graphpad.com)
Procedure
文章信息
版权信息
© 2020 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Besserer-Offroy, E., Brouillette, R. L., Longpré, J. and Sarret, P. (2020). Assessing Gαq/15-signaling with IP-One: Single Plate Transfection and Assay Protocol for Cell-Based High-Throughput Assay. Bio-protocol 10(16): e3715. DOI: 10.21769/BioProtoc.3715.
- Luginina, A., Gusach, A., Marin, E., Mishin, A., Brouillette, R., Popov, P., Shiriaeva, A., Besserer-Offroy, E., Longpre, J. M., Lyapina, E., Ishchenko, A., Patel, N., Polovinkin, V., Safronova, N., Bogorodskiy, A., Edelweiss, E., Hu, H., Weierstall, U., Liu, W., Batyuk, A., Gordeliy, V., Han, G. W., Sarret, P., Katritch, V., Borshchevskiy, V. and Cherezov, V. (2019). Structure-based mechanism of cysteinyl leukotriene receptor inhibition by antiasthmatic drugs. Sci Adv 5(10): eaax2518.
分类
神经科学 > 细胞机理 > 胞内信号传导
神经科学 > 细胞机理 > 受体-配体结合
细胞生物学 > 细胞信号传导 > 胞内信号传导
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link