- EN - English
- CN - 中文
Methylation-sensitive Amplified Polymorphism as a Tool to Analyze Wild Potato Hybrids
利用甲基化敏感扩增多态性分析野生马铃薯杂交种
发布: 2020年07月05日第10卷第13期 DOI: 10.21769/BioProtoc.3671 浏览次数: 5221
评审: Samik BhattacharyaYing FengAnonymous reviewer(s)
Abstract
Methylation-Sensitive Amplification Polymorphism (MSAP) is a versatile marker for analyzing DNA methylation patterns in non-model species. The implementation of this technique does not require a reference genome and makes it possible to determine the methylation status of hundreds of anonymous loci distributed throughout the genome. In addition, the inheritance of specific methylation patterns can be studied. Here, we present a protocol for analyzing DNA methylation patterns through MSAP markers in potato interspecific hybrids and their parental genotypes.
Keywords: MSAP (甲基敏感扩增多态性)Background
Nucleotide sequences are not the only form of genomic information: DNA methylation, histone proteins, enzymes that modify histones and nucleotide residues on DNA and even RNAs, influence gene activity and provide another layer of instructions to the cell. Epigenetic changes, also called epimutations, can be inherited and have important phenotypic consequences. In plants, methylation reactions modify cytosine residues into 5-methylcytosines. This epigenetic mechanism is essential to maintain the genomic integrity and contributes to regulate gene expression in developmental processes and in response to biotic and abiotic stresses. In addition, changes in DNA methylation are triggered by genomic shocks like hybridization and polyploidization, two vital phenomena in plant evolution (Cara et al., 2019).
There are different alternatives to study changes in DNA methylation. Global cytosine methylation can be assessed by using high-performance liquid chromatography (HPLC), analytical methods that allow to quantify cytosines and 5-methylcytosines and to calculate the percentage of methylated residues in the genome. For studying the DNA methylation at specific positions on the genome two alternatives can be mentioned. One is to use isoschizomers with different sensitivities to methylation at cytosines of the restriction site. For example, Methylation-Sensitive Amplification Polymorphism (MSAP) markers characterize the methylation pattern at anonymous 5′-CCGG sequences from random genomic DNA. This is an adaptation of the original AFLP protocol (Vos et al., 1995) substituting the frequent cutter enzyme MseI by HpaII and MspI. These enzymes recognize the same tetranucleotide restriction site (5′-CCGG), but HpaII is sensitive to full methylation (both strands methylated) and cleaves the hemi-methylated external cytosine, whereas MspI is sensitive only to methylation of the external cytosines of the restriction site. Another possibility to study site-specific methylation status is by using a bisulphite sequencing methodology. Bisulphite treatments on DNA convert cytosines into uracils while 5-methylcytosines remain unchanged. Then, by sequencing amplicons (i.e., a target gene or promoter) of bisulphited and control DNA, it is possible to distinguish between methylated and unmethylated cytosines. With the development of next generation sequencing technologies, whole-genome bisulphite sequencing, or WGBS, can be implemented to infer the position for all 5-methylcytosines in a genome. However, this approach requires a high-quality reference genome to perform epigenetic analyses. Although the increasing number of draft genomes and the reduction in sequencing costs offer possibilities to implement massive methylation analyses in non-model species, the use of MSAP markers continues to be a valuable tool in many laboratories.
Wild potatoes (Solanum, section Petota) are a group of species related to the cultivated potato Solanum tuberosum L. Internal breeding barriers can be incomplete, thus, interspecific hybridization occurs in areas of sympatry (Camadro et al., 2012). Epigenetic changes in response to interspecific hybridization have been documented in synthetic and natural hybrids of wild potato species (Cara et al., 2019). Solanum x rechei H. & H. is a hybrid species that grows in sympatry with its wild progenitors, Solanum kurtzianum B. & W. and Solanum microdontum B. Here, we present a protocol for analyzing DNA methylation patterns through MSAP markers in hybrids and their parental genotypes. Using an R script, fragments present in the synthetic hybrids are categorized as S. microdontum or S. kurtzianum species-specific if they are present on the parental genotypes, S. x rechei species-specific, if they are present in at least one of the S. x rechei evaluated genotypes or as novel if they are only observed in the synthetic hybrids.
Materials and Reagents
- Consumables
- Chemicals
- Tris base (BIOPACK, catalog number: 200016 6800)
- Ethylenediaminetetraacetic acid (EDTA) (BIOPACK, catalog number: 2000964500 )
- Sodium Chloride (BIOPACK, catalog number: 200016 4606)
- Cetyltrimethyl ammonium bromide (CTAB) (Bio Basic INC, catalog number: DB0108 )
- β-mercaptoethanol (BIOPACK, catalog number: 2000954500 )
- Chloroform (BIOPACK, catalog number: 200016 5100)
- Isoamyl alcohol (BIOPACK, catalog number: 2000972500 )
- Ethyl alcohol (BIOPACK, catalog number: 200016 5400)
- Sodium acetate (BIOPACK, catalog number: 200016 8000)
- RNase A (Thermo Fisher Scientific, catalog number: EN0531 )
- Lambda DNA/EcoRI+HindIII Marker (Promega, catalog number: G1731 )
- Glycerol (BIOPACK, catalog number: 200016 2000)
- Bromophenol blue (BIOPACK, catalog number: 2000962200 )
- Xylene cyanol FF (SIGMA, catalog number: X4126 )
- Boric acid (BIOPACK, catalog number: 2000935900 )
- Agarose (TransGen, catalog number: GS201 )
- UltraPureTM Ethidium Bromide (Thermo Fisher Scientific, catalog number: 15585011 )
- EcoRI (New England Biolabs, catalog number: R0101S )
- Bovine Serum Albumin (BSA) (Promega, catalog number: R3961 )
- HpaII (New England Biolabs, catalog number: R0171S )
- MspI (New England Biolabs, catalog number: R0106 )
- T4 DNA Ligase (Promega, catalog number: M1801 )
- Oligonucleotides (Table 1) (IDT, Integrated DNA Technologies, Inc., Iowa, USA)
Table 1. Sequences of adaptors and primers used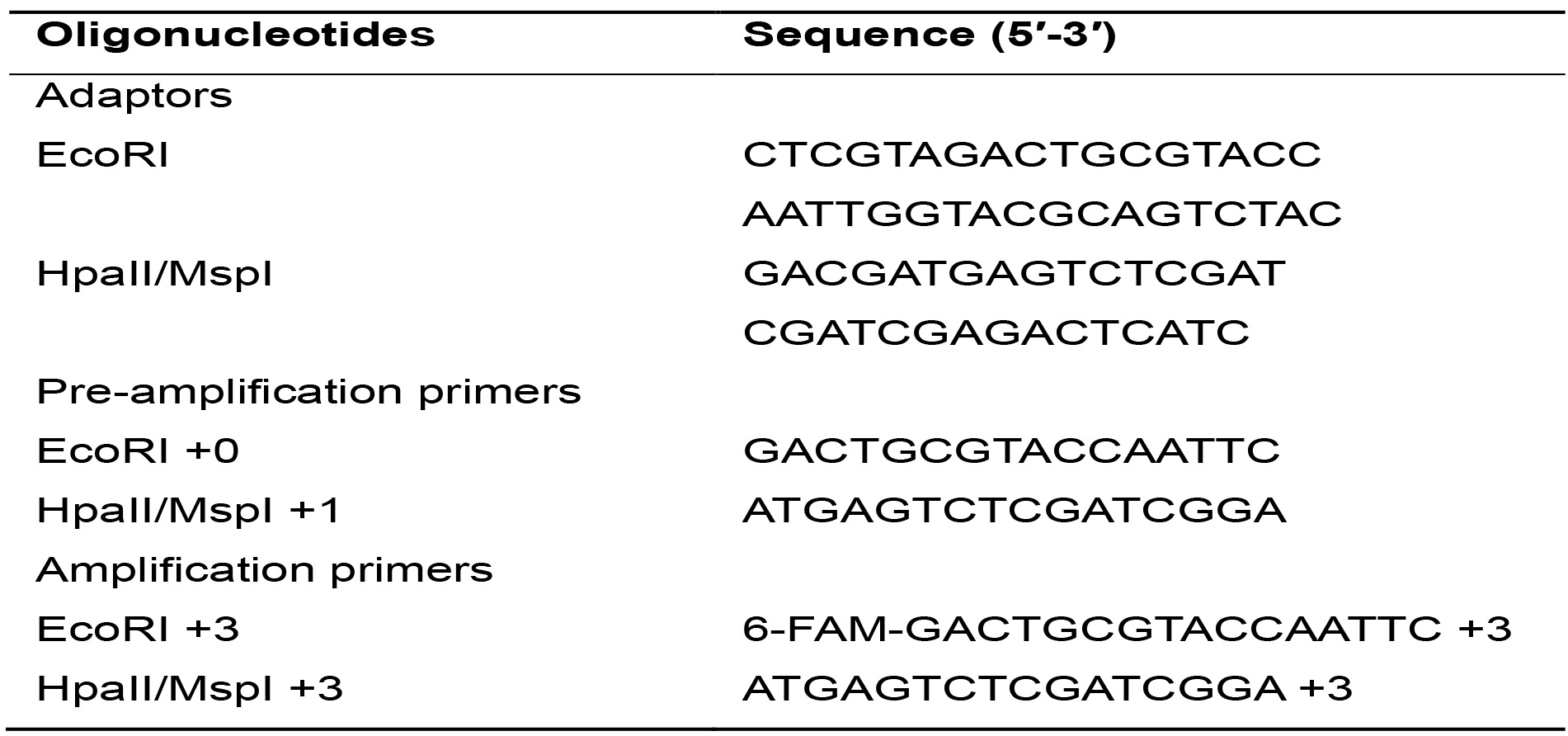
- Taq DNA Polymerase (Thermo Fisher Scientific, catalog number: 11615044 )
- MgCl2 (Thermo Fisher Scientific, catalog number: 11615044 )
- dNTP Set (100 mM) (Thermo Fisher Scientific, catalog number: 10297018 )
- Hi-Di formamide (Thermo Fisher Scientific, catalog number: 4311320 )
- GeneScan 500HD Rox size standard (Thermo Fisher Scientific, catalog number: 401734 )
- ddH2O (sterile)
- Extraction buffer (see Recipes)
- TBE (Tris-Borate-EDTA) buffer (see Recipes)
- 6x DNA loading buffer (see Recipes)
Equipment
- Pipettes (BOECO, Wheaton SOCOREX and Finnpipette)
- Vortex (IKA, model: MS1 )
- UltraCentrifuge (Eppendorf, model: 5804R )
- Thermostatic bath (Vicking, model: Masson )
- Agarose gel electrophoresis system (Bio-Rad Laboratories, model: 1645056 )
- Microwave oven (HITPLUS, model: CM203M )
- Spectrophotometer (AmpliQuant, model: AQ-07 )
- Gel Imager (Bio-Rad Laboratories, model: Gel Doc 1000 )
- Incubator (SANYO, model: MIR 262 )
- Thermocycler Veriti 96 well (Applied Biosystems, model: 9902 )
- Genetic Analyzer (Invitrogen, model: ABI PRISM 3130 )
- Ultra-low Freezer -80 °C (Forma Scientific, model: 8270 )
Software
- GeneMapper v3.7. (Applied Biosystems, Foster City, CA, USA)
- R v2.15.1. (R Foundation for Statistical Computing, Vienna, Austria)
- RStudio v1.1.442 (RStudio Inc., Boston, MA, USA)
Procedure
文章信息
版权信息
© 2020 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Cara, N., Marfil, C. F., Bertoldi, M. V. and Masuelli, R. W. (2020). Methylation-sensitive Amplified Polymorphism as a Tool to Analyze Wild Potato Hybrids. Bio-protocol 10(13): e3671. DOI: 10.21769/BioProtoc.3671.
分类
植物科学 > 植物分子生物学 > 遗传分析
分子生物学 > DNA > 基因分型
分子生物学 > DNA > 电泳
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link