- EN - English
- CN - 中文
High-throughput Site-directed Scanning Mutagenesis Using a Two-fragment PCR Approach
利用双片段PCR法进行高通量定点扫描突变
(*contributed equally to this work) 发布: 2020年01月05日第10卷第1期 DOI: 10.21769/BioProtoc.3484 浏览次数: 6616
评审: Pooja VermaVinay PanwarAnonymous reviewer(s)
Abstract
Site-directed scanning mutagenesis is a useful tool applied in studying protein function and designing proteins with new properties, such as increased stability or enzymatic activity. Creating a systematic library of hundreds of site-directed mutants is still a demanding and expensive task. The established protocols for making such libraries include PCR amplification of the recombinant DNA using a pair of primers carrying a target mutation in the same PCR. Unfortunately, this approach is very often coupled with PCR artifacts which compromise overall efficiency of site-directed mutagenesis. To reduce the failure rate due to the PCR artifacts, we have set up a high-throughput mutagenesis protocol based on a two-fragment PCR approach. To this end, each mutation is introduced in two separate PCRs resulting in two linear fragments of the mutated plasmid. In the next steps, the PCR template is digested and the two matching plasmid fragments are joined together using Gibson assembly. Separating the corresponding mutagenic primers into two different PCRs decreases a number of artifacts and thus increases overall cloning efficiency. Furthermore, free software that we developed facilitates both high-throughput primer design and analysis of sequencing results. Overall, this protocol enabled us to efficiently produce several alanine-scanning libraries of 400 single-point mutations with complete coverage of the protein sequence.
Keywords: Scanning mutagenesis (扫描诱变)Background
High-throughput cloning techniques are widely used for research involving expression and purification of proteins. Site-directed scanning mutagenesis techniques in particular are often used to generate mutants for functional studies or to generate stabilized proteins for biophysics and structural biology. Libraries necessary for these purposes may contain several hundred mutants and their generation can be expensive and time consuming. Many single-point mutant libraries have been created using single vector PCRs with overlapping primers, e.g. QuikChange (Agilent). Although this and similar mutagenesis techniques are well established and widely used (for a recent review see Ortega et al., 2019), they may lead to a number of different errors in the final vector (Liu and Naismith, 2008; Edelheit et al., 2009; Sun et al., 2013; Heydenreich et al., 2017) caused by inherent sequence overlap of primers. Such errors include additional mutations and primer repeats, which necessitates sequencing of multiple colonies or additional rounds of mutagenesis. The protocol presented here is based on a two-fragment PCR mutagenesis, where the primers used in each PCR reaction have no sequence complementarity. The two halves of a plasmid to be cloned are amplified in two separate PCRs, followed by in vitro circularisation using Gibson assembly reaction (Figure 1). As a result, the number of successful mutagenesis reactions after sequencing only one clone per mutant is increased, allowing us to generate multiple alanine-scanning libraries of complete proteins of ca. 400 amino acids in length within approx. 6 weeks each (Heydenreich et al., 2017). In addition, our protocol can also be used to insert multiple point mutations in one round of cloning and for sequence modification (e.g., addition of purification tags, small insertions and deletions) with increased efficiency.
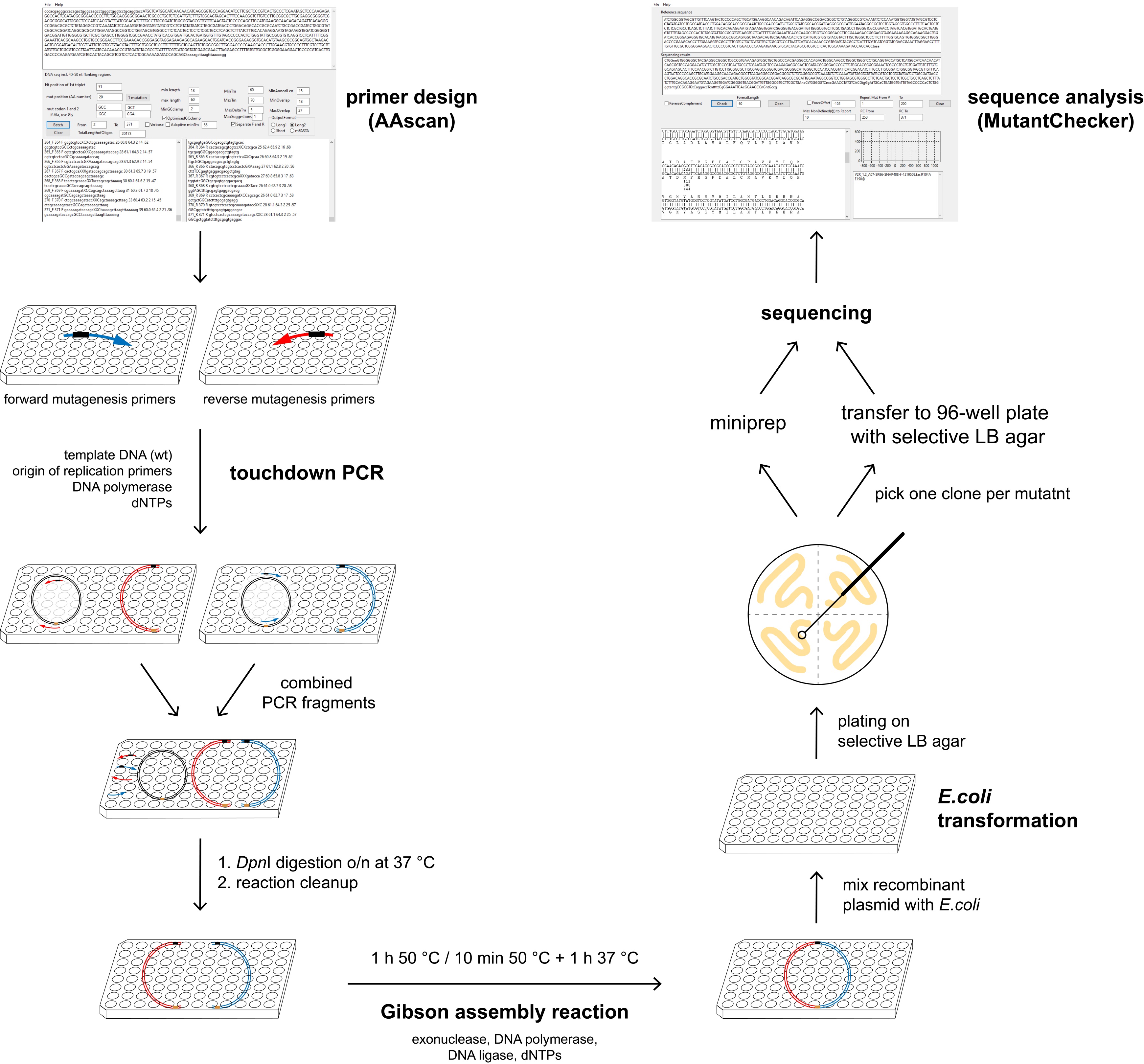
Figure 1. Overview of the mutagenesis technique. AAscan software (Sun et al., 2013) is used to design mutagenic primers for the high-throughput site-directed scanning mutagenesis. Two PCR reactions are done per mutant, in each of them approximately half of the vector is amplified. Two fragments containing one mutation are combined, followed by DpnI digestion at 37 °C overnight. Reaction clean-up is performed to purify DNA fragments, which are then assembled by Gibson assembly reaction. Bacteria are transformed with the resulting circular plasmid and plated on selective LB agar plates. One clone per mutant is sent for sequencing either on a selective LB agar 96-well plate or as purified DNA. All steps excluding the plating of the bacteria are done in 96-well plates. Sequencing results are analyzed in a high-throughput manner using MutantChecker program (Sun et al., 2013).
Materials and Reagents
Note: All reagents must be of analytical grade or higher and stored according to recommendations from the manufacturer. All plasticware must be DNase-free.
- LB-agar 96-well plates with the appropriate antibiotic(s) (can be ordered from a sequencing company; if not, see Recipes)
- Pipette tips, 10 μl, 200 μl and 1,250 μl (VWR, European catalog numbers: 613-1093, 613-1092 and 613-1087)
- (Optional) Sterile toothpicks
- Polypropylene conical tubes 15 ml and 50 ml (Falcon, catalog numbers: 352096 and 352070)
- Microcentrifuge tubes 1.5 ml (Simport, catalog number: T330-7G)
- Pipetting reservoir (Diversified Biotech, catalog number: REBP-3000)
- 96-well PCR plates, 0.2 ml (VWR, European catalog number: 211-0297)
- Microseal ’B’ PCR plate seals (Bio-Rad, catalog number: MSB1001)
- Deep-well plates 96/2,000 μl (Eppendorf, catalog number: 651033405)
- Adhesive gas-permeable film for culture plates (VWR, European catalog number: 391-1261)
- Sterile Petri dishes (e.g., ThermoScientific NuclonTM Delta Surface, catalog number 150350)
- Disposible sterile spreaders (e.g., VWR, catalog number: 612-5497) or autoclaved glass beads 2 mm (e.g., Supelco, catalog number: 1.04014)
- ZR-96 DNA Clean & Concentrator (Zymo Research, catalog number: D4024)
- Chemically competent Escherichia coli XL-1 Blue cells (Agilent, catalog number: 200249) or some other cell strain which is used for cloning (Mach1, DH5α, TOP10 etc.)
- Cloning primers
ColE1A: 5′-GGAGCGAACGACCTACACCGAACTGAGATACCTACAGCG-3′
ColE1B: 5′-CGCTGTAGGTATCTCAGTTCGGTGTAGGTCGTTCGCTCC-3′
Note: This pair of primers can be used for all plasmids containing the ColE1 origin of replication. - Plasmid to be used as a template in PCR
- Nicotinamide adenine dinucleotide, NAD+, 50 mM (New England Biolabs, catalog number: B9007L)
- Deoxynucleotide (dNTPs) solution mix, 10 mM each dNTP (New England Biolabs, catalog number: N0447S)
- T5 exonuclease, 10 U/μl (New England Biolabs, catalog number: M0363S)
- Phusion HF DNA polymerase, 2 U/μl (New England Biolabs, catalog number: M0530L)
- Taq DNA ligase, 40 U/μl (New England Biolabs, catalog number: M0208L)
- DpnI, 20 U/μl (New England Biolabs, catalog number: R0176L)
- Nuclease-free water (Cell Signaling Technology, catalog number: 12931S)
- Phusion High-Fidelity PCR Master Mix with GC Buffer (New England Biolabs, catalog number: M0532L) or some other appropriate DNA polymerase master mix (see Procedure B. PCR)
- PEG-8000 (Sigma-Aldrich, catalog number: P5413)
- Tris-HCl buffer, 1 M, pH 7.5 (Thermo Fischer Scientific, catalog number: 15567027)
- MgCl2, 1 M (Sigma-Aldrich, catalog number: M1028)
- DL-dithiothreitol (DTT), 1 M (Sigma-Aldrich, catalog number: 43816)
- SOC outgrowth medium (New England Biolabs, catalog number: B9020S)
- Antibiotic stock solution(s) (depending on an antibiotic resistance gene(s) carried by the plasmid of interest)
- Lysogeny broth (LB) broth with agar (Sigma-Aldrich, catalog number: L2897)
- Ultrapure deionized water with electrical resistivity of 18 MΩ cm at 25 °C
- 5x isothermal reaction (IT) buffer (see Recipes)
- 1.33x Gibson assembly mixture (see Recipes)
- LB-agar plates with the appropriate antibiotic(s)-Petri dishes (see Recipes)
Equipment
- Diverse pipettes including multi-channel pipettes and repeat pipettors
- Thermocycler that can accommodate 96-well PCR plates (e.g., EppendorfTM MastercyclerTM Nexus Thermal Cycler, catalog number: E6332000029)
- Centrifuge for deepwell plates (e.g., Eppendorf 5810 R with swing-bucket rotor A-4-62, catalog number: 5811000320)
- Laboratory incubator set at 37 °C
- Laboratory incubator/shaker set at 37 °C
Software
- AAscan (Sun et al., 2013; https://github.com/dmitryveprintsev/AAScan/)
- MutantChecker (Sun et al., 2013; https://github.com/dmitryveprintsev/AAScan/)
Procedure
文章信息
版权信息
© 2020 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Heydenreich, F. M., Miljuš, T., Milić, D. and Veprintsev, D. B. (2020). High-throughput Site-directed Scanning Mutagenesis Using a Two-fragment PCR Approach. Bio-protocol 10(1): e3484. DOI: 10.21769/BioProtoc.3484.
分类
分子生物学 > DNA > DNA 克隆
分子生物学 > DNA > 诱/突变
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link