- EN - English
- CN - 中文
Lysosome Targeting RedGreen-assay: Selective Autophagy Sensing Assay for Mammalian Cells
溶酶体靶向红绿试验
(*contributed equally to this work) 发布: 2019年12月20日第9卷第24期 DOI: 10.21769/BioProtoc.3455 浏览次数: 5943
评审: Alka MehraMarco Di GioiaAlexandros C Kokotos

相关实验方案

研究免疫调控血管功能的新实验方法:小鼠主动脉与T淋巴细胞或巨噬细胞的共培养
Taylor C. Kress [...] Eric J. Belin de Chantemèle
2025年09月05日 3543 阅读
Abstract
The process of autophagy is an essential cellular mechanism, required to maintain general cell health through the removal of dysfunctional organelles, such as the ER, peroxisomes and mitochondria, as well as protein aggregates, and bacteria. Autophagy is an extremely dynamic process, and tools are constantly being developed to study the various steps of this process. This protocol details a method to study the end steps of autophagy-lysosomal fusion and the formation of the autolysosome. Many techniques have been used to study the various steps of the autophagy process. Here we describe the RedGreen-assay (RG-assay), an immunofluorescence-based technique used to visualize the targeting of substrates to the autolysosome in live cells. This technique takes advantage of the low lysosomal pH and over-expression of a tandem GFP-mCherry tagged protein targeted to an organelle of interest. While in the neutral cytosol or autophagosome, both GFP and RFP will fluoresce. However, within the autolysosome, the GFP signal is quenched due to the low pH environment and the RFP emission signal will predominate. This technique is readily quantifiable and amenable to high throughput experiments. Additionally, by tagging the GFP-RFP tandem fluorescent protein with organelle specific targeting sequences, it can be used to measure a wide range of substrates of autophagy.
Keywords: Autophagy (自噬)Background
Macroautophagy (herein referred to as autophagy) is an essential cellular process, required for the removal of large cellular material. It can be both general and selective, either removing bulk material or specific cytoplasmic material, respectively. The method described in this protocol can be used in the study of either process, but the focus here will be on selective autophagy. In brief, selective autophagy in mammalian cells begins with the tagging of damaged organelles or other cellular material, generally with the small protein ubiquitin. The collection of ubiquitinated proteins on the substrate allows the recruitment of autophagy receptors which interact with the ubiquitin protein and with the ATG8 family of autophagic factors on developing phagophores through their LC3 interacting region (LIR). This interaction results in the engulfment of the cargo by the phagophore, producing an autophagosome. Finally, the autophagosome fuses with the lysosome, producing an autolysosome and degrading the cargo within. Selective autophagy is responsible for degrading cargo such as peroxisomes (pexophagy), mitochondria (mitophagy), protein aggregates (aggrephagy) and bacteria (xenophagy). For a review, refer to Lahiri et al., 2019 and Morishita and Mizushima, 2019. As autophagy proceeds in many steps, different assays have been developed to study the various stages. Historically, the most popular assay to detect changes in autophagy induction is the activation of the ATG8 family of proteins, particularly LC3B. Another common assay for detecting targeting of substrate to the autolysosome is the co-localization between the cargo with either the autophagosome marker LC3, or with various lysosomal markers such as Lamp1. Co-localization studies are technically easy to perform, can be performed in a high throughout manner and can detect specific cargo within the lysosome easily through the use of fluorescent tagging. However, it is limited by the microscope resolution, and the cargo can be difficult to detect due to degradation. For a review describing these and other methods for detecting autophagy, refer to Klionsky et al., 2016.
The protocol described here, lysosome targeting Red-Green (RG) assay, can be used to study the end stages of autophagy, namely the localization of the cargo to the autolysosome. This assay takes advantage of the acidic pH of the lysosome (~4.6) and its ability to quench GFP (pKa ~6) fluorescence without affecting RFP (pKa = 4.5) fluorescence. Through the use of a tandem RFP-GFP fusion protein targeted to an organelle of choice, cargo that is cytosolic or within an autophagosome, and therefore exposed to neutral pH, will appear yellow when using pseudo colors green for GFP and red for RFP due to the fluorescence of both GFP and RFP. Upon lysosomal fusion, the labeled cargo will be exposed to an acidic pH, quenching GFP fluorescence and resulting in mainly RFP fluorescence (Figure 1). Again, this can be detected by microscopy and the ratio of cargo with RFP-only fluorescence can be compared to the total RFP fluorescence, to obtain a measure of autophagy. This method was first used to study aggrephagy (Pankiv et al., 2007). By tagging LC3 with a tandem GFP-mCherry, autophagic vesicles and autolysosomes were able to be readily visualized and distinguished. We have adopted the principle used with LC3 to visualize the targeting of peroxisomes to autolysosomes. This is done by tagging the peroxisome targeting transmembrane domain of PEX26 with the tandem fluorophore, allowing it to be localized to the cytosolic face of the peroxisomal membrane (Deosaran et al., 2013; Riccio et al., 2019). This allows for the visualization of pexophagy specifically. Others have also used a similar technique to study pexophagy, where the fluorescent proteins were targeted to the peroxisome matrix through the use of a matrix targeting sequence (Nazarko et al., 2014). Importantly, our method, targeting the fluorophore to the cytosolic face of the peroxisome membrane, allows for increased sensitivity of the tandem fluorophore to the acidic environment of the lysosome. Additionally, our quantification method differs, taking into account the relative GFP and mCherry fluorescence in order to determine the quantity of peroxisomes targeted to the lysosome. In addition to our use of the tandem fluorophore in pexophagy, we and others have used this method to quantify mitophagy (Allen et al., 2013; Kim et al., 2014; Wang et al., 2015).
We have shown that the use of the tandem fluorophore is a reproducible method for observing and quantifying selective autophagy. As displayed above, the tandem mCherry-GFP lysosome targeting assay is extremely flexible, and can be used to measure various forms of selective autophagy, as well as general autophagy. Through the pH sensitivity of the fluorophores, it is a reliable method to detect lysosomal targeting. Additionally, because GFP and mCherry are expressed at equal levels, comparing the fluorescent intensities of mCherry to GFP allows for a reproducible method to quantify pexophagy.
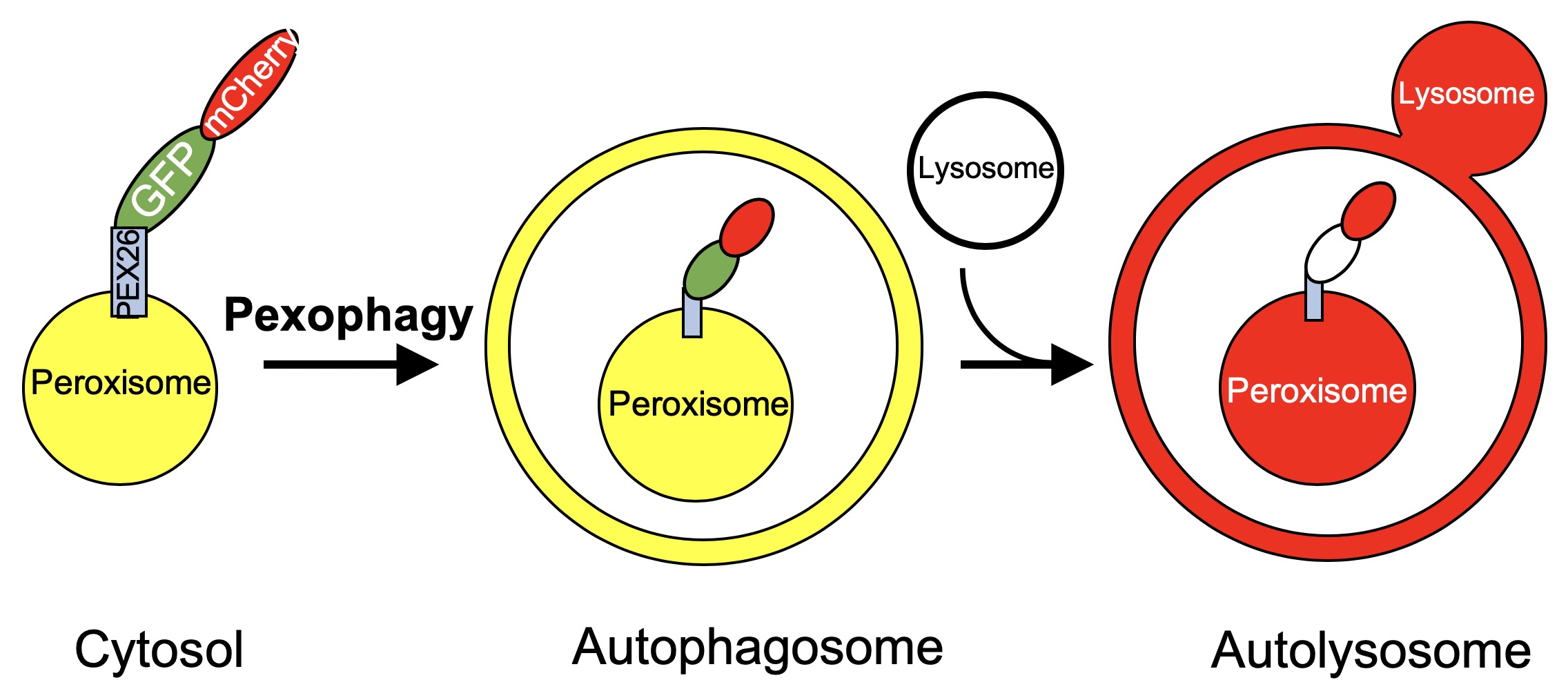
Figure 1. Schematic of the lysosome targeting RG assay. Peroxisomes expressing the PEX26TM-RG protein will appear yellow within the cytosol or the autophagosome due to fluorescence of both GFP and mCherry. When targeted to the lysosome, GFP fluorescence will be quenched and the peroxisome will appear red.
Materials and Reagents
- Stericup Quick Release Millipore Express PLUS 0.22 μm PES filter, 500 ml (Millipore Sigma, catalog number: S2GPU05RE)
- T25 flask (Corning Life Sciences, catalog number: C353109)
- 4-well NuncTM Lab-TekTM Chambered Coverglass (Thermo Fisher Scientific, catalog number: 155383)
- Lipofectamine 2000 (Life Technologies, catalog number: 11668-019)
- E-64 (Enzo Life Sciences, catalog number: BML-PI107-0001)
- Leupeptin (Bioshop, catalog number: LEU001)
- HeLa human cervical cancer cell line (ATCC CCL-2)
- GFP-mCherry (RG)-PEX26TM plasmid
- Dulbecco’s Modified Eagle’s Medium (Hyclone, catalog number: SH3008101)
- L-Glutamine (Hyclone, catalog number: SH30034.01)
- Fetal Bovine Serum (Life Technologies, catalog number: A12617)
- OptiMEM Reduced Serum Medium (Thermo Fisher, catalog number: 31985062)
- Trypsin-EDTA (0.05%), phenol red (Thermo Fisher, catalog number: 25300062)
- CO2 independent medium (Life Technologies, catalog number: 18045)
- Hank’s Balanced Salt Solution (HBSS) (Thermo Fisher, catalog number: 14025092)
Equipment
- Zeiss LSM710 equipped with a 63x 1.4 NA oil immersion objective and the appropriate lasers (488 nm on a tuneable Argon 458/477/488/514 nm at 30 mM with a 493-565 nm bandpass filter and 561 nm DPSS laser with a 600-700 nm bandpass)
- Forma Steri-Cycle CO2 Incubator (Thermo Fisher)
Software
- Volocity (Perkin Elmer, version 6.3)
- Zeiss Zen (Zeiss, https://www.zeiss.com/microscopy/int/products/microscope-software/zen.html)
- Microsoft® Office Excel (Microsoft Office 365)
Procedure
文章信息
版权信息
© 2019 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Riccio, V., Vissa, M., McQuibban, A. and Kim, P. K. (2019). Lysosome Targeting RedGreen-assay: Selective Autophagy Sensing Assay for Mammalian Cells. Bio-protocol 9(24): e3455. DOI: 10.21769/BioProtoc.3455.
- Riccio, V., Demers, N., Hua, R., Vissa, M., Cheng, D. T., Strilchuk, A. W., Wang, Y., McQuibban, G. A. and Kim, P. K. (2019). Deubiquitinating enzyme USP30 maintains basal peroxisome abundance by regulating pexophagy. J Cell Biol 218(3): 798-807.
分类
免疫学 > 免疫细胞功能 > 巨噬细胞
微生物学 > 微生物-宿主相互作用 > 细菌
神经科学 > 细胞机理 > 突触生理学
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link