- EN - English
- CN - 中文
Proximity Ligation Assay for the Investigation of the Intramolecular Interaction of ELMO1
邻位连接技术用于ELMO1分子内相互作用研究
(*contributed equally to this work) 发布: 2019年12月05日第9卷第23期 DOI: 10.21769/BioProtoc.3449 浏览次数: 4779
评审: Edgar Soria-GomezTongxing SongAnonymous reviewer(s)

相关实验方案
Cell-Sonar:通过特定蛋白标志物表达变化追踪目标蛋白的简便低成本方法
Sabrina Brockmöller [...] Simone Rothmiller
2025年02月05日 1655 阅读
Abstract
Intramolecular interaction is a common mechanism that regulates protein activities. Conventionally, such interactions are investigated by classical in vitro biochemical assays. Here, we describe a protocol for studying the intramolecular interaction of cell motility and engulfment 1 (ELMO1) in mammalian cells by using proximity ligation assay (PLA). PLA is a specific and sensitive method that allows the observation of interacting proteins by target-specific antibody detection coupled to rolling circle amplification. ELMO1 is the regulatory subunit of ELMO1-dedicator of cytokinesis 180 (DOCK180) bipartite Rac1 guanine nucleotide exchange factor (GEF) which adopts a closed autoinhibitory conformation via an intramolecular interaction of its N-terminal ELMO inhibitory domain (EID) and C-terminal ELMO autoregulatory domain (EAD). In the assay, PLA signals are detected in cells transfected with ELMO11-315 and ELMO1315-727 fragments. Moreover, overexpression of FE65, a neuronal adaptor which has been shown to disrupt ELMO1 intramolecular interaction, reduces the PLA signals of the two ELMO1 fragments significantly. Together, our results demonstrate that PLA can be employed for studying protein intramolecular interaction.
Keywords: Autoinhibition (自身抑制)Background
The activities of proteins in cells can be regulated by various mechanisms including intramolecular interaction (Miernyk and Thelen, 2008). Several protein-protein interaction assays have been adopted for studying intramolecular interaction such as co-immunoprecipitation and protein complementation assays (PCAs). Despite being widely employed, there are known limitations of these conventional assays. For instance, co-immunoprecipitation is not ideal for detecting low-affinity interaction. Even though PCAs provide good sensitivity, the reporter fragments may hinder the interaction of interest (Ohad et al., 2007; Miernyk and Thelen, 2008). Proximity ligation assay (PLA) is an innovative technology that allows in situ detection of protein-protein interactions. The protein targets are first labeled by their specific primary antibodies from different species, followed by the recognition of species-specific secondary antibodies conjugated to oligonucleotides (PLA probes). Proximity ligation of the oligonucleotides generates the templates for single-stranded rolling circle amplification. The resultant products are detected with fluorescence-labeled complementary oligonucleotide detection probes. PLA offers exceptional specificity and sensitivity as target-specific antibodies and single-stranded rolling circle amplification are involved. The resultant products are detected with fluorescence-labeled complementary oligonucleotide detection probes, and PLA signals can be visualized and quantified by using fluorescence microscopy (Weibrecht et al., 2010). Although the technology has been available for over a decade, the possibility of using PLA for investigating protein intramolecular interaction has not been explored.
Engulfment and cell motility 1 (ELMO1) is the regulatory subunit of the bipartite ELMO1-dedicator of cytokinesis 180 (DOCK180) guanine nucleotide exchange factor (GEF) for the activation of the small GTPase Rac1. ELMO1 possesses several distinct domains: a Ras binding domain (RBD), ELMO inhibitory domain (EID), ELMO domain, pleckstrin homology (PH) domain, ELMO autoregulatory domain (EAD) and a PXXP motif (Figure 1). Similar to many other GEFs, the activity of ELMO1 is modulated by the intramolecular interaction between its EID and EAD which allows ELMO1 to adopt a closed autoinhibitory conformation at the basal state (Patel et al., 2010) (Figure 1). The relief of ELMO1 autoinhibition is required for the targeting of the complex of ELMO1-DOCK180 to the plasma membrane where it stimulates Rac1 (Patel et al., 2011). However, the mechanism that alleviates the autoinhibition of ELMO1 remains elusive. We have recently reported that the neuronal adaptor protein FE65 interacts with ELMO1 EAD and to disrupt the interaction of ELMO11-315 (comprises of ELMO1 amino acid residues 1-315) and ELMO1315-727 (comprises of ELMO1 amino acid residues 315-727) fragments, which possess the EID and EAD respectively, by using both classical protein binding assay and PLA (Li et al., 2018). Therefore, we demonstrate for the first time that PLA can be applied for examining protein intramolecular interaction.
Here, we describe how to investigate the intramolecular interaction of ELMO1 EID and EAD by using PLA. The method may also be adopted for the investigation of other protein intramolecular interactions.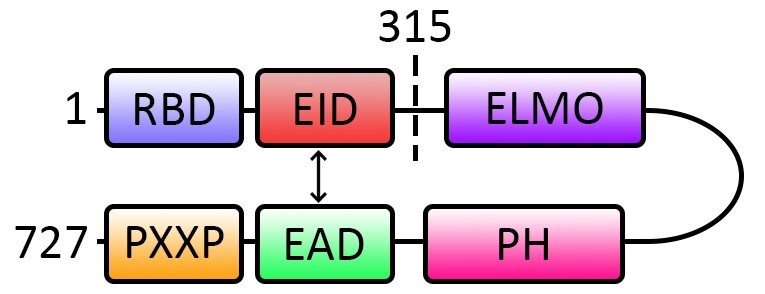
Figure 1. Schematic diagram shows the subdomains and the intramolecular interaction of ELMO1. At basal stage, the EAD and EID interact intramolecularly (indicated with double headed arrow) to form an autoinhibitory conformation. The disruption of the intramolecular interaction alleviates the autoinhibition for the subsequence Rac1 activation. Numbers denote the amino acid residue number in ELMO1. RBD: Ras-binding domain; EID: ELMO inhibitory domain; ELMO: ELMO domain; PH: Pleckstrin homology domain; EAD: ELMO autoregulatory domain; PXXP: PXXP motif.
Materials and Reagents
- SterilinTM Standard 90 mm Petri Dish (Thermo Scientific, catalog number: 101VIRR), stored at room temperature
- 12-well cell culture plate (SPL, catalog number: 30012), stored at room temperature
- Cover glass 18-mm circles (Thermo-Menzel, catalog number: CB00180RA1), stored at room temperature
- Hemocytometer (HBG, catalog number: 9010-01), stored at room temperature
- Microscope slides (Thermo Scientific, catalog number: 6776214), stored at room temperature
- HEK293 cells (ATCC, catalog number: CRL-1573), stored in liquid nitrogen
- Goat anti-ELMO1 antibody (Santa Cruz Biotechnology, catalog number: sc-21651), stored at 4 °C
- Mouse anti-FLAG antibody (Sigma-Aldrich, catalog number: F1804), stored at -20 °C
- Rabbit anti-β-tubulin antibody (Abcam, catalog number: AB6046)
- Donkey anti-rabbit IgG (H + L) highly cross-adsorbed secondary antibody, Alexa Fluor 488 (Invitrogen, catalog number: A21206), stored at 4 °C
- Fetal bovine serum (HyClone, catalog number: SV30160.03), stored at -20 °C
- pCMV-Tag2B vector (Aligent, catalog number: 211172), stored at -20 °C
- T4 DNA Ligase (1 U/μl) (Thermo Scientific, catalog number: 15224025), stored at -20 °C
- BamHI (10 U/μl) (Thermo Scientific, catalog number: ER0055), stored at -20 °C
- XhoI (10 U/μl) (Thermo Scientific, catalog number: ER0691), stored at -20 °C
- TIANprep Mini Plasmid Kit (TIANGEN, catalog number: DP103), stored at room temperature
- QuikChange II Site-Directed Mutagenesis Kit (Agilent, catalog number: 200523), stored at -20 °C
- Phusion High-Fidelity PCR Kit (Thermo Scientific, catalog number: F553S), stored at -20 °C
- UltraPureTM Agarose (Invitrogen, catalog number: 16500500), stored at room temperature
- TAE Buffer (Tris-acetate-EDTA) (50x) (Thermo Scientific, catalog number: B49), stored at room temperature
- UltraPureTM Ethidium Bromide, 10 mg/ml (Invitrogen, catalog number: 15585011), stored at room temperature
- 1 kb Plus DNA Ladder (New England Biolabs, catalog number: N3200), stored at -20 °C
- Luria Broth Base, powder (Invitrogen, catalog number: 12795084), stored at room temperature
- Clear nail polish
- LB Agar, powder (Invitrogen, catalog number: 22700041), stored at room temperature
- Ampicillin sodium salt (Sigma-Aldrich, catalog number: A0166), stored at 4 °C
- Kanamycin sulfate (Sigma-Aldrich, catalog number: 60615), stored at 4 °C
- Poly-D-lysine hydrobromide (Sigma-Aldrich, catalog number: P7280), stored at -20 °C
- DMEM (HyClone, catalog number: SH30021.01), stored at 4 °C
- Trypsin-EDTA 0.05% (Gibco, catalog number: 25300062), stored at -20 °C
- Opti-MEM reduced serum medium (Gibco, catalog number: 31985088), stored at 4 °C
- X-tremeGENETM 9 DNA transfection reagent (Sigma-Aldrich, catalog number: 6365779001), stored at 4 °C
- PBS (Gibco, catalog number: 10010023), stored at room temperature
- Paraformaldehyde (Sigma-Aldrich, catalog number: P6148), stored at 4 °C
- Triton X-100 (Anatrace, catalog number: T1001500ML), stored at room temperature
- Duolink® In Situ Red Starter Kit Mouse/Goat (Sigma-Aldrich, catalog number: DUO92103), stored at -20 °C
- 1x TAE buffer (see Recipes)
- Poly-D-lysine solution (see Recipes)
- 4% paraformaldehyde (freshly prepared) (see Recipes)
- 1x Ligation buffer (see Recipes)
- 1x Amplification red (see Recipes)
- Wash buffer A (see Recipes)
- Wash buffer B (see Recipes)
Equipment
- Water bath (Labnet, model: W1106)
- Heat block (Labnet, model: D1301-230V)
- UV transilluminator (Accuris, model: E3000)
- T100TM Thermal Cycler (Bio-rad)
- HerathermTM Compact Microbiological Incubators (37 °C, Thermo Scientific)
- FormaTM Series II 3110 Water-Jacketed CO2 Incubators (37 °C, 5% CO2, Thermo Scientific)
- Fluorescence microscope (Nikon, model: Eclipse Ni-U) equipped with QI2 high-resolution microscope camera (Nikon)
Software
- NIS-Elements research basic (Nikon)
- Prism (https://www.graphpad.com/scientific-software/prism/)
Procedure
文章信息
版权信息
© 2019 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Chan, W. W. R., Chau, D. L. D., Li, W. and Lau, K. (2019). Proximity Ligation Assay for the Investigation of the Intramolecular Interaction of ELMO1. Bio-protocol 9(23): e3449. DOI: 10.21769/BioProtoc.3449.
- Li, W., Tam, K. M. V., Chan, W. W. R., Koon, A. C., Ngo, J. C. K., Chan, H. Y. E. and Lau, K. F. (2018). Neuronal adaptor FE65 stimulates Rac1-mediated neurite outgrowth by recruiting and activating ELMO1. J Biol Chem 293(20): 7674-7688.
分类
生物化学 > 蛋白质 > 相互作用 > 蛋白质-蛋白质相互作用
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link