- EN - English
- CN - 中文
Imaging VIPER-labeled Cellular Proteins by Correlative Light and Electron Microscopy
利用关联光学和电子显微镜技术进行VIPER标记细胞蛋白成像
发布: 2019年11月05日第9卷第21期 DOI: 10.21769/BioProtoc.3414 浏览次数: 6650
Abstract
Advances in fluorescence microscopy (FM), electron microscopy (EM), and correlative light and EM (CLEM) offer unprecedented opportunities for studying diverse proteins and nanostructures involved in fundamental cell biology. It is now possible to visualize and quantify the spatial organization of cellular proteins and other macromolecules by FM, EM, and CLEM. However, tagging and tracking cellular proteins across size scales is restricted by the scarcity of methods for attaching appropriate reporter chemistries to target proteins. Namely, there are few genetic tags compatible with EM. To overcome these issues we developed Versatile Interacting Peptide (VIP) tags, genetically-encoded peptide tags that can be used to image proteins by fluorescence and EM. VIPER, a VIP tag, can be used to label cellular proteins with bright, photo-stable fluorophores for FM or electron-dense nanoparticles for EM. In this Bio-Protocol, we provide an instructional guide for implementing VIPER for imaging a cell-surface receptor by CLEM. This protocol is complemented by two other Bio-Protocols outlining the use of VIPER (Doh et al., 2019a and 2019b).
Keywords: Protein tag (蛋白标签)Background
Multiple protein targets can be imaged at once by fluorescence microscopy (FM), electron microscopy (EM), or correlative light and EM (CLEM) (Giepmans et al., 2005; Lucas et al., 2012; Philimonenko et al., 2014; Johnson et al., 2015; Kim et al., 2015). FM enables multi-color microscopy in both living and fixed cells, and acquiring data can be relatively fast and easy. However, EM offers better resolution for imaging nanoscale features, including cell receptors, membrane boundaries, neuronal connections (Hildebrand et al., 2017), chromatin organization (Ou et al., 2017), or the endocytic machinery (Sochacki et al., 2017). We anticipate an increased reliance on multi-color, cross-platform imaging for investigating proteins associated with normal cell function and human diseases (Megason and Fraser, 2007; Lichtman et al., 2008; Milne and Subramaniam, 2009; Muller and Heilemann, 2013; Plaza et al., 2014; Kremer et al., 2015; Lucocq et al., 2015; Karreman et al., 2016; Laine et al., 2016; Romero-Brey and Bartenschlager, 2017). Currently, most high-resolution imaging studies obtain protein-specific contrast with immunolabeling, which has known shortcomings (Berglund et al., 2008; Bordeaux et al., 2010; Baker, 2015; Bradbury and Pluckthun, 2015).
The central obstacle that has limited progress in multi-scale microscopy is the shortage of genetic tags for labeling proteins. There are a number of protein tags available for imaging proteins by FM, including fluorescent proteins and self-labeling enzyme tags (Sunbul and Yin, 2009; Hinner and Johnsson, 2010). However, there are few genetic tags for EM or CLEM (Ellisman et al., 2012). Most EM tags rely on the oxidation of diaminobenzidine (DAB) to form a polymer that is stained with osmium tetroxide to generate contrast. Examples include APEX (Martell et al., 2012; Lam et al., 2015), miniSOG (Shu et al., 2011), the tetracysteine tag (Gaietta et al., 2002), and others (Kuipers et al., 2015; Liss et al., 2015). Among the DAB-reliant EM tags, miniSOG, FLIPPER (Kuipers et al., 2015), and the tetracysteine tag are compatible with CLEM. However, DAB staining is finicky, and it can be difficult to localize the stain sufficiently to resolve targets. Further progress in multi-scale microscopy will require new methods for labeling proteins with EM- and CLEM-compatible reporters, such as quantum dots (Qdots) (Giepmans et al., 2005) or FluoroNanogoldTM particles (Takizawa et al., 2015).
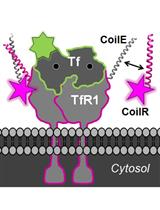
We recently described a new class of tags for multiscale microscopy called Versatile Interacting Peptide (VIP) tags (Tane et al., 2017; Doh et al., 2018). VIP tags use heterodimerizing coiled-coil peptides to label proteins. One coil is expressed as a fusion to the protein of interest. This coil has a partner, the probe peptide, that is conjugated to a reporter molecule to deliver protein-specific contrast. The probe peptide can be conjugated to a number of reporters, such as fluorophores, small molecules (e.g., biotin), or nanoparticles.
In this Bio-Protocol, we outline the use of VIPER to image a transmembrane receptor by CLEM. VIPER is a VIP tag comprised of CoilE tag and CoilR probe peptide. In Procedure A, we describe the plating and transfection of cells to express a CoilE-tagged receptor: transferrin receptor 1 (TfR1-CoilE). Procedure B describes how to label a cell receptor with CoilR-biotin for subsequent detection with streptavidin-Qdot655. In Procedure C we have an illustrated guide on how to mount ITO coverslips to a slide holder for correlative fluorescence imaging. Procedure D describes image acquisition with a commercially available CLEM microscope, the FEI CorrSightTM. In Procedures E and F we describe methods for preparing samples for EM. Procedure G details the acquisition of SEM micrographs on a Helios NanolabTM 660 EM. We additionally developed a quantitative image analysis pipeline for automated image segmentation on high magnification SEM images (see the Data Analysis section). This program runs in Matlab and reports the number of nanoparticles within a field of view in SEM micrographs.
This protocol is published in tandem with two supporting publications detailing the use of VIPER (Figure 1). Doh et al. (2019a) details how to generate CoilR probe peptides, including biotinylated CoilR. Doh et al. (2019b) describes imaging VIPER-labeled proteins in cells by FM.
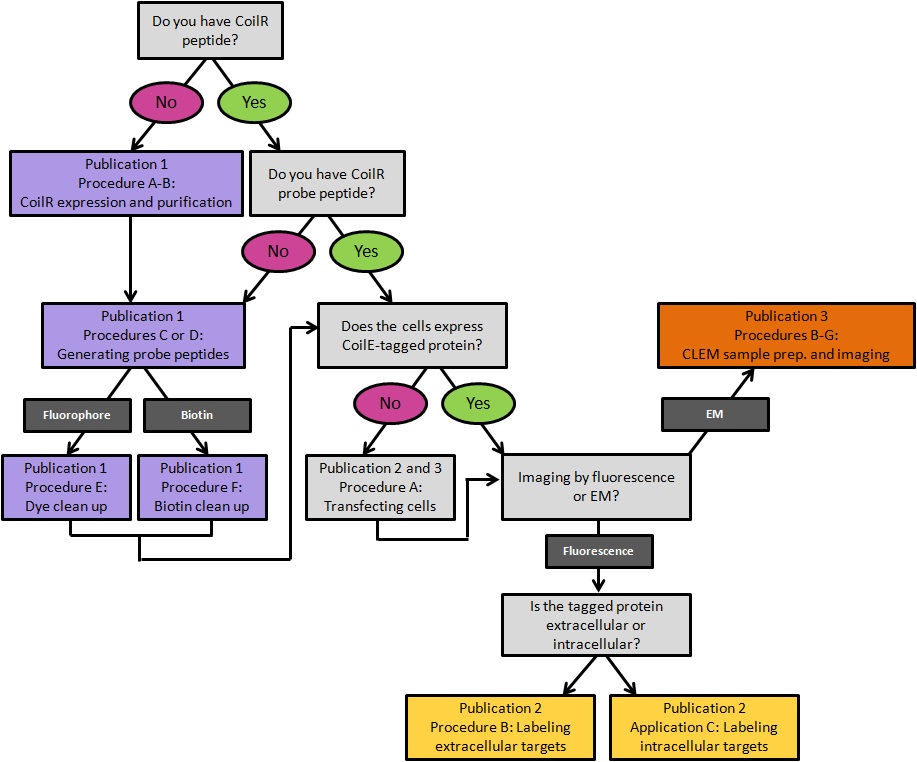
Figure 1. A decision tree for implementing VIPER for labeling cellular proteins. Procedures are color-coded by the publication in which they appear. Publication 1: Doh et al., 2019a; Publication 2: Doh et al., 2019b; Publication 3: this article.
Materials and Reagents
Note: “*” indicates a brand that is critical to the success of the experiment.
Materials
- Aluminum coverslip holder
Note: We used a custom machined aluminum plate with a hole for a 22 x 22 mm coverslip. This plate is 76 x 26 x 1.5 mm with a 12 mm diameter hole in the center. - *Indium tin-oxide 22 x 22 mm coverslips (2SPI, catalog number: 06486-AB)
- Tape (Scotch® MagicTM Tape)
- Desiccator cabinet (Thermo Scientific NalgeneTM, catalog number: 53170070)
- Desiccant (DrieriteTM, catalog number: D1085)
- Kimwipes (Kimtech, catalog number: 34120)
- *Conductive silver paint “Leitsilber” (Ted Pella, catalog number: 16035)
- *SEM pin stub specimen mount (Ted Pella, catalog number: 16144)
- *Carbon thread (Leica, catalog number: 16771511116)
- LDPE 500 ml squeeze wash bottle (Thermo Scientific, catalog number: 24010500)
- Transfer bulb pipette (VWR, catalog number: 16001-182)
- CHO TRVb cells (courtesy of Prof. Timothy McGraw, Cornell University, Ithaca, New York) (McGraw et al., 1987)
- Ham’s F-12 Medium (Life Technologies, GibcoTM, catalog number: 11765062)
- Dulbecco’s phosphate-buffered saline without calcium or magnesium; DPBS (GibcoTM, catalog number: 14190144)
- Trypsin-EDTA (0.25%) (Life Technologies, GibcoTM, catalog number: 25200056)
- Fetal bovine serum (FBS) (GE, HycloneTM, catalog number: SH30910.03)
- *LipofectamineTM 2000 (Thermo Scientific, catalog number: 11668019)
- Opti-MEM (Life Technologies, GibcoTM, catalog number: 31985070)
- *Streptavidin-QdotTM 655 conjugate (Invitrogen, catalog number: Q10121MP)
- Anhydrous ethanol (Decon Labs, catalog number: 2716)
- 20% (v/v) paraformaldehyde (PFA) stock (Electron Microscopy Sciences, catalog number: 15713S)
- Live Cell Block solution (see Recipes)
- Qdot Block solution (see Recipes)
- Qdot Labeling solution (see Recipes)
Equipment
- Hemocytometer (Hausser Scientific, catalog number: 1475)
- Humidified CO2 Incubator (New Brunswick Galaxy 170S, catalog number: C0170S-120-0000)
- Tissue culture hood (Thermo Scientific, model: 1300 Series A2)
- Tissue culture inverted light microscope (Carl Zeiss, Zeiss Primovert)
- Fine point tweezers (Ted Pella Dumostar Biology, catalog number: 525-PS)
- Diamond-tipped scribe (Ted Pella, catalog number: 54468)
- Stainless steel crinkle washers (Tousimis Washers, catalog number: 8767-01)
- Orbital Shaker (Stovall Belly DancerTM, catalog number: BDRAA1158)
- Spinning disk confocal fluorescence microscope (FEI CorrSightTM)
- 63x objective lens (Carl Zeiss, 1.4 NA Plan-Apochromat M27, catalog number: 420780-9900-000)
- 5x objective lens (Carl Zeiss, 0.16 NA EC Plan-Neofluar M27, catalog number: 420330-9901-000)
- Scanning electron microscope (FEI Helios NanolabTM 660 SEM)
- Critical point dryer (Leica EM CPD300)
- High Vacuum Flash Carbon Coating Machine (Leica EM ACE600)
Software
- FEI MAPS (FEI version 2.1.38.1199 and version 3.0)
- FEI Helios NanolabTM xT Microscope Control (FEI version 5.5.1 and version 10.1.7)
- Matlab (Mathworks Software Version R2017b)
Procedure
文章信息
版权信息
© 2019 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Doh, J. K., Chang, Y. H., Enns, C. A., Lόpez, C. S. and Beatty, K. E. (2019). Imaging VIPER-labeled Cellular Proteins by Correlative Light and Electron Microscopy. Bio-protocol 9(21): e3414. DOI: 10.21769/BioProtoc.3414.
分类
癌症生物学 > 增殖信号转导 > 细胞生物学试验 > 化学抗性
细胞生物学 > 细胞成像 > 荧光
生物化学 > 蛋白质 > 标记
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link