- EN - English
- CN - 中文
SIRF: A Single-cell Assay for in situ Protein Interaction with Nascent DNA Replication Forks
SIRF:一种蛋白与新生DNA复制叉原位相互作用的单细胞检测办法
发布: 2019年09月20日第9卷第18期 DOI: 10.21769/BioProtoc.3377 浏览次数: 8043
评审: Gal HaimovichAnonymous reviewer(s)
Abstract
The duplication of DNA is a fundamental process that is required for the transfer of the genetic information from parent to daughter cells. Aberrant DNA replication processes are associated with diverse disease phenotypes, including developmental defects, ageing disorders, blood disorders such as Fanconi Anemia, increased inflammation and cancer. Therefore, the development of tools to study proteins associated with error-free DNA replication processes is of paramount importance. So far, methods to study proteins associated with nascent replication forks relied on conventional immunofluorescence and immunoprecipitation assays of 5′-ethylene-2′-deoxyuridine (EdU) labeled DNA (iPOND). While greatly informative and important, these methods lack specificities for nascent fork interactions (e.g., IF) or assay an average change of millions of cells without single-cell resolution (e.g., iPOND). The assay system described here combines proximity ligation assay (PLA) with EdU coupled click-iT chemistry, which we termed “in situ Protein Interaction with Nascent DNA Replication Forks (SIRF)”. This method enables sensitive and quantitative analysis of protein interactions with nascent DNA replication forks with single-cell resolution, and can further be paired with conventional immunofluorescence marker analysis for added multi-parameter analysis.
Keywords: SIRF (SIRF)Background
Defects in high fidelity DNA replication are associated with severe biological outcomes. These amongst others include developmental abnormalities, premature aging, hematological disorder and cancer (Loeb and Monnat, 2008; Magdalou et al., 2014). To avoid DNA replication instability, cells have evolved intricate mechanisms to ensure error-free propagation of its genetic material. There is an ever-growing list of disease-suppressing proteins that actively ensure DNA replication stability and function at all stages of DNA replication processes. A comprehensive understanding of the functions of DNA replication stability proteins is imperative to allow development of effective and specific agents targeting disease outcomes.
Historically, visualization of proteins localized at or near DNA replication forks was advanced by the advent of nucleoside analogs, such as 5-bromo-2’-deoxyuridine (BrdU) that is incorporated into newly replicating DNA and further detected with fluorescent-labeled antibodies (Gonchoroff et al., 1986; Leif et al., 2004). Immunofluorescence (IF) assays examining co-localization of proteins with the BrdU-label indicates that the protein is present in close vicinity to actively replicating DNA (Sengupta et al., 2003). This method provided critical information on replication processes. However, it is an indirect method that lacks the resolution that would be required to unequivocally distinguish protein-DNA interactions from proteins located near DNA. Another ground-breaking procedure was iPOND (standing for isolation of proteins on nascent DNA) that allows to detect and isolate proteins on newly replicated DNA (Petermann et al., 2010; Sirbu et al., 2011 and 2013; Dungrawala et al., 2015). In this procedure, nascent DNA is labeled with the thymidine analog EdU, which is then biotinylated using click-IT chemistry (Moses and Moorhouse, 2007). The newly synthesized biotinylated DNA is immuno-precipitated with streptavidin-coated agarose or magnetic beads. This so isolates proteins that bind to newly replicated DNA, which either can be detected by Western blotting or by mass spectrometry. While this method revolutionized the DNA replication field by enabling to directly study protein interactions with replication forks, iPOND detects the average of thousands of replication forks and cells. iPOND lacks the single-cell level resolution that is achieved by IF, which can provide valuable additional parameter information such as stage of the S-phase, or cell identity. Moreover, the technique is labor-intensive and requires copious amounts of starting material (~100,000,000 cells per condition). The assay system described here termed in situ protein interaction with nascent DNA replication forks (SIRF) (Roy et al., 2018a and 2018b) combines EdU-biotin Click-chemistry with proximity ligation assay (PLA) to overcome these challenges. The PLA technology developed by Soderberg et al. for single-molecule protein-protein interaction studies, utilizes antibodies with oligonucleotides conjugates, that can be ligated into a circular DNA molecule when two antibodies are in close proximity to each other (< 40 nm) (Soderberg et al., 2008). Once ligated, the DNA circle can be amplified by rolling circle DNA polymerase. After annealing of fluorescent DNA probes that are complementary to the DNA circle, this procedure results in a highly amplified fluorescent signal, so that even single molecule interactions are readily detectable by conventional immunofluorescence microscopy. In SIRF, PLA is used with antibodies against biotin (detecting biotinylated EdU) and a protein of interest, which results in a PLA signal only if the protein is located within 40 nm or less from the newly synthesized DNA (Figure 1). The SIRF method requires little amounts of cells (~100-1,000), preserves the single-cell resolution as seen with IF, and can be readily and accurately quantified. Moreover, the single-cell resolution provides additional valuable information including cell morphology, cell-cycle stage (early or late S-phase), spatial orientation of protein signal in the nucleus, which can be associated with changes in replication fork-protein dynamics. The SIRF assay furthermore can be combined with conventional IF for additional cell-biomarker analysis, which amongst others allows to examine replication proteins in a heterogeneous cell population that can be differentiated by lineage specific markers. In this case, primary and secondary antibody staining for IF is performed prior to the PLA reaction to avoid interference of PLA secondary antibodies from cross-reacting with IF primary antibodies. The SIRF assay can be used to study protein dynamics at ongoing and stalled DNA replication forks, as well as chromatin maturation at newly replicated DNA regions (Petruk et al., 2017, Roy et al., 2018a and 2018b).
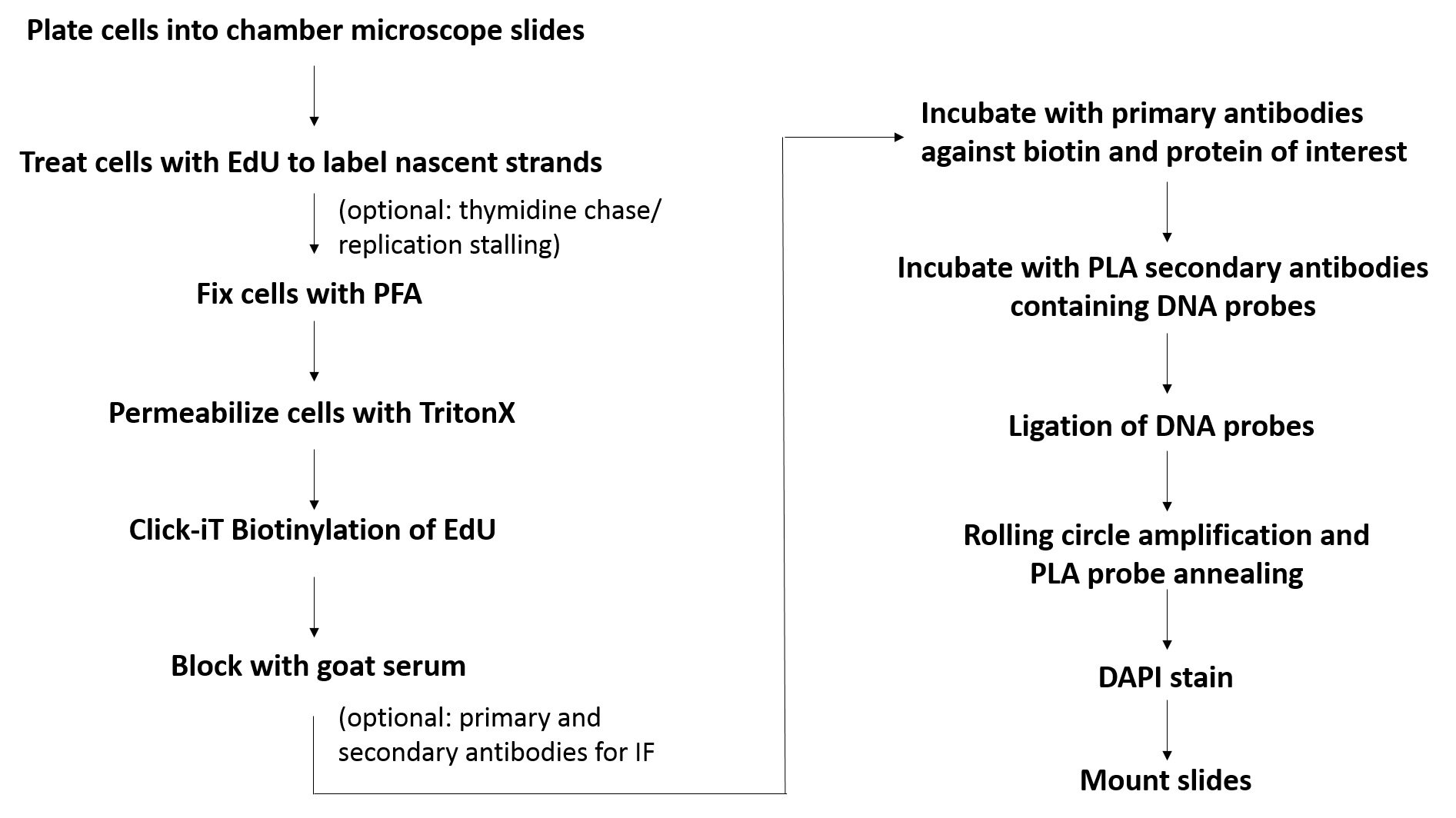
Figure 1. Flowchart for the SIRF assay. Shown is the basic workflow and the major steps involved in SIRF assay.
Materials and Reagents
- 8-well chamber microscope slides (Thermo Scientific, Nunc, catalog number: 177402)
- Plastic cover slips (Electron Microscopy Sciences, catalog number: 72261-50)
- Glass cover slips (Fisher Scientific, catalog number: 12-548-5M)
- Kimwipes (Fisher Scientific, catalog number: NC9855580)
- Aluminum foil (Fisher Scientific, catalog number: 01-213-105)
- Paper towel (Envision, catalog number: 23304)
- 0.22 µm bottletop filter (Corning, catalog number: 430758)
- Thymidine (Sigma-Aldrich, catalog number: T1895)
- Rabbit anti-biotin antibody (Cell Signaling, catalog number: 5597S)
- Mouse anti-biotin antibody (Sigma-Aldrich, catalog number: B7653
- Rabbit anti-RPA antibody (Abcam, catalog number: ab79398)
- Goat serum (Sigma-Aldrich, catalog number: G9023)
- 5′-ethylene-2′-deoxyuridine (EdU) (Invitrogen, catalog number: A10044)
- Hydroxyurea (HU) (Sigma-Aldrich, catalog number: H8627)
- Paraformaldehyde 32% solution, EM grade (EMS, catalog number: 15714)
- Triton X-100 (Sigma-Aldrich, catalog number: T8787)
- Biotin Azide (Invitrogen, catalog number: B10184)
- Alexa Fluor 488 azide (Invitrogen, catalog number: A10266)
- Copper sulfate solution (Fluka Analytical, catalog number: 35185)
- Sodium ascorbate (Sigma-Aldrich, catalog number: 11140)
- Phosphate buffered saline (PBS) (Sigma-Aldrich, catalog number: P4417)
- Duolink® Mouse plus PLA probe (Sigma-Aldrich, catalog number: DUO92001-100RXN)
- Duolink® Rabbit minus PLA probe (Sigma-Aldrich, catalog number: DUO92005-100RXN)
- Duolink® PLA detection reagent red (Sigma-Aldrich, catalog number: DUO92008-100RXN)
- Duolink® Duolink In Situ Wash Buffers (Sigma-Aldrich, catalog number: DUO82049-20L)
- 4’, 6-diamidino-2-phenylindole (DAPI) (Life Technologies, catalog number: 62248)
- Prolong Gold antifade reagent (Invitrogen, catalog number: P36934)
- Tween 20 (Sigma-Aldrich, catalog number P7949)
- Hydrochloric acid (Fisher Scientific, catalog number: A144-212)
- Tris base (Fisher Scientific, catalog number: BP152-1)
- IMDM (Gibco, catalog number: 12440-053)
- FBS (Gemini Bio-products, catalog number: 100-106)
- Sodium Chloride (Fisher Scientific, S271-3)
- Fixation solution (see Recipes)
- Permeabilization solution (see Recipes)
- Blocking Buffer (see Recipes)
- Wash Buffer A (see Recipes)
- Wash Buffer B (see Recipes)
Equipment
- Autoflow IR water jacketed CO2 incubator (NUAIRE, model: NU-4750)
- Nikon eclipse Ti-U inverted microscope (Nikon, model: Ti-U)
- Coplin jar (Thermo ScientificTM E94)
- Fine curved forceps (Fine Science Tools, Dumont #7, catalog number: 11271-30)
- Slide box (VWR, catalog number: 82003-414)
- Vortexer (VWR analog vortex mixer, catalog number: 10153-838)
- 4 °C refrigerator (BSI, model: SCGP21OW1AREF)
- -20 °C freezer (BSI, model: ABT-2020MB)
Software
- Duolink® Analysis tool
(Sigma-Aldrich, http://www. Sigma-aldrich. com/ catalog/ product/ sigma / duo90806? Lang = en & ion = US) - NIS-elements (Nikon, https://www.nikoninstruments.com/Products/Software)
- Microsoft Excel (Microsoft, https://products.office.com/en-us/excel)
- ImageJ (ImageJ, https://imagej.net/Welcome)
- GraphPad Prism (GraphPad, https://www.graphpad.com/scientific-software/prism/)
Procedure
文章信息
版权信息
© 2019 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Roy, S. and Schlacher, K. (2019). SIRF: A Single-cell Assay for in situ Protein Interaction with Nascent DNA Replication Forks. Bio-protocol 9(18): e3377. DOI: 10.21769/BioProtoc.3377.
- Roy, S., Luzwick, J. W. and Schlacher, K. (2018a). SIRF: Quantitative in situ analysis of protein interactions at DNA replication forks. J Cell Biol 217(4): 1521-1536.
分类
癌症生物学 > 基因组不稳定性及突变 > 细胞生物学试验 > DNA结构和改变
细胞生物学 > 细胞染色 > 核酸
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link