- EN - English
- CN - 中文
Photoaffinity Labeling of Respiratory Complex I in Bovine Heart Submitochondrial Particles by Photoreactive [125I] amilorides
利用光敏阿米洛利光亲和标记牛心脏亚线粒体颗粒中呼吸链复合物I
发布: 2019年09月05日第9卷第17期 DOI: 10.21769/BioProtoc.3349 浏览次数: 4712
评审: Alexandros AlexandratosAlexander GalkinAnonymous reviewer(s)
Abstract
The architecture of quinone/inhibitor-access channel in proton-translocating NADH-quinone oxidoreductase (respiratory complex I) was modeled by X-ray crystallography and cryo-EM, however, it remains debatable whether the channel model reflects the physiologically relevant state present throughout the catalytic cycle. Using photoreactive [125I]amilorides, we demonstrated that amiloride-type inhibitors bind to the interfacial region of multiple subunits (49-kDa, ND1, PSST, and 39-kDa subunits), which is difficult to reconcile with the current channel model. This report describes the procedures for photoaffinity labeling of bovine submitochondrial particles by photoreactive [125I]amilorides. The protocol could be widely applicable for the characterization of various biologically active compounds, whose target protein remains to be identified or characterized.
Keywords: Photoaffinity labeling (光亲和标记)Background
Proton-translocating NADH-quinone oxidoreductase (respiratory complex I) is a multi-subunit membrane protein complex, which catalyzes the initial step of mitochondrial/bacterial electron transport chains (Hirst, 2013). The recent progress in X-ray crystallography and cryo-EM enabled modeling the entire structure of complex I. The structural studies proposed the quinone/inhibitor-access channel model to explain how quinone/inhibitor bind to the enzyme (Sazanov, 2015; Wirth et al., 2016). However, since there is currently no structural data of complex I with bound-quinone or inhibitor, it remains debatable whether the channel model reflects the physiologically relevant state present throughout the catalytic cycle.
Photoaffinity labeling technique, which uses a synthetic ligand that possesses photoreactive group such as diazirine and phenyl-azido, is the most commonly used method of affinity-based protein modification (Hatanaka and Sadakane, 2002). It provides a powerful means of investigating interactions between biologically active compounds and the proteins of interest. Recently, to get insights into the structure of quinone/inhibitor-binding site in mitochondrial complex I, we carried out photoaffinity labeling using photoreactive [125I]amilorides ([125I]PRA3, [125I]PRA4, [125I]PRA5, and [125I]PRA6) with bovine heart submitochondrial particles (SMPs) (Uno et al., 2019). The radioisotope (125I) was used as a tracer, which enables the experiment with the lowest concentrations of ligands (1-10 nM). These concentrations are approximately two orders of magnitude lower than those of alkyne-tagged amilorides (PRA1 and PRA2, Murai et al., 2015), which can be conjugated with biotin or fluorophores via Cu+-catalyzed click chemistry (Wang et al., 2003) after cross-linking reaction. This advantage may minimize the possibility of non-specific labeling, which is a primary cause of false-positive results.
We herein describe the detailed procedures of the photoaffinity labeling of complex I in bovine SMPs by [125I]PRA5 as an example (Figure 1), which includes 1) preparation of 125I-tagged photoreactive ligand, 2) cross-linking of bovine SMPs by the ligand, and 3) identification of the labeled protein(s).
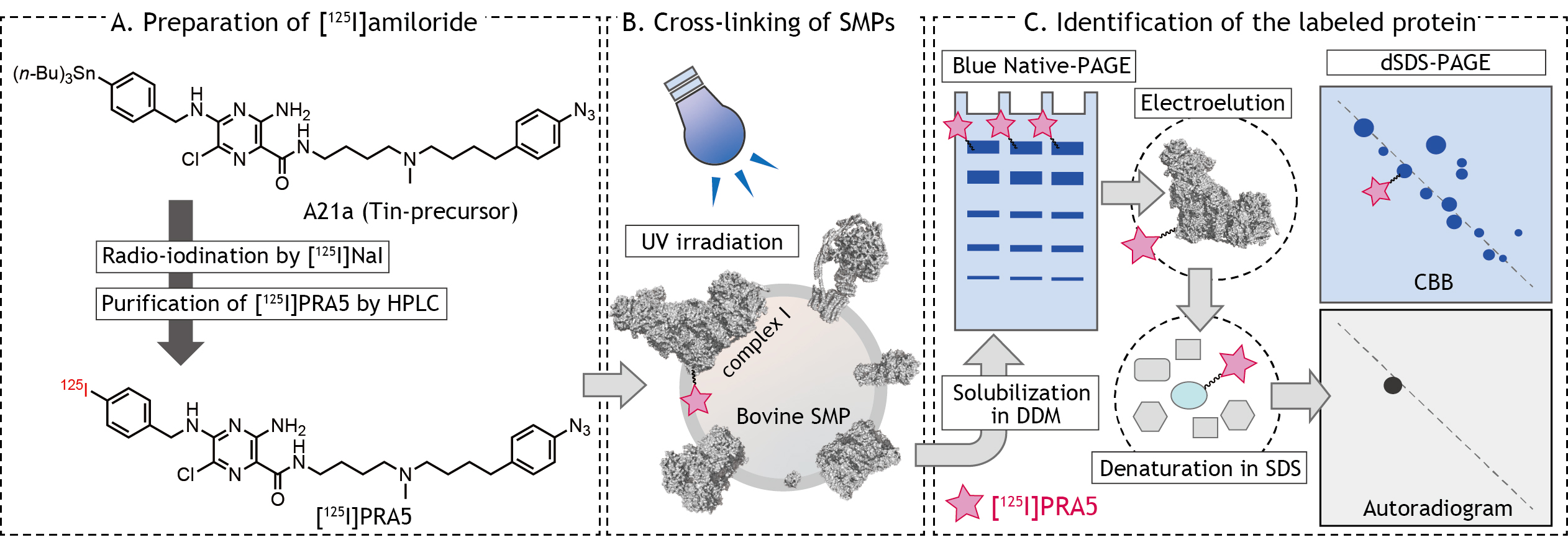
Figure 1. Schematic presentation of photoaffinity labeling of bovine heart SMPs by [125I]amilorides
Materials and Reagents
- 1.5 ml Screw-capped microtube (Watson, catalog number: 139-115-112C)
- 1.5 ml microtube (Safe-Lock Tube, Eppendorf, catalog number: 0030120086)
- Pipette tips (Gilson)
- Membrane filter unit (Amicon-Ultra, 100 kDa, Merck Millipore, catalog number: UFC510096)
- Reverse-phase HPLC column (COSMOSIL 5C18-MSII, 4.6 mm x 200 mm, Nacalai Tesque)
- TLC plate (Merck Millipore, catalog number: 1.05554.0001)
- Plastic wrap
- Amiloride-tin-precursor (A21a, 1.0 mM in ethanol, Figure 1A, The synthetic procedures are described in Uno et al., 2019)
- [125I]NaI (carrier-free, 2,000 Ci/mmol, Perkin-Elmer, catalog number: NEZ033A)
- Chloramine T (Wako Pure Chemicals, catalog number: 032-02182)
- Sodium hydrogen sulfite (NaHSO3, Wako Pure Chemicals, catalog number: 198-01371)
- Bovine submitochondrial particles (SMPs)
Notes:- SMPs are inside-out vesicles of inner mitochondrial membrane.
- Mitochondria were isolated from bovine heart as described elsewhere (Smith, 1967).
- SMPs were prepared on the basis of the previously described protocol (Matsuno-Yagi and Hatefi, 1985) using a sonication medium containing 0.25 M sucrose, 1.0 mM potassium succinate, 1.5 mM ATP, 10 mM MgCl2, 10 mM MnCl2, and 10 mM Tris/HCl (pH 7.4), followed by ultracentrifugation. They are stored in a buffer containing 250 mM sucrose and 10 mM Tris-HCl (pH 7.4) at -80 °C until used.
- NativePAGETM Bis-Tris protein gels (4-16% precast gel, Thermo Fisher, catalog number: BN1004BOX)
- NativePAGETM Runnning buffer and Cathode buffer additive (Thermo Fisher, catalog numbers: BN2001 and BN2002, respectively)
- Serva Blue G (CBB G-250, Serva, catalog number: 35050.01)
- n-Dodecyl-D-β-maltoside (DDM, Dojin, catalog number: D316)
- Glycerol (Wako Pure Chemicals, catalog number: 075-00616)
- Ethanol (Wako Pure Chemicals, catalog number: 057-00456)
- Methanol (Wako Pure Chemicals, catalog number: 137-01823)
- Acetic acid (Wako Pure Chemicals, catalog number: 017-00251)
- Sucrose (Wako Pure Chemicals, catalog number: 192-00017)
- MgCl2 (hexahydrate, Wako Pure Chemicals, catalog number: 135-00165)
- NaCl (Wako Pure Chemicals, catalog number: 195-01665)
- Ponceau S (Nacalai Tesque, catalog number: 28322-72)
- Tricine (Dojin, catalog number: GB05)
- SDS (Wako Pure Chemicals, catalog number: 191-07145)
- Mercaptethanol (Wako Pure Chemicals, catalog number: 137-06862)
- Tris (Wako Pure Chemicals, catalog number: 011-20095)
- Acrylamide (Wako Pure Chemicals, catalog number: 011-08015)
- Bis-acrylamide (Wako Pure Chemicals, catalog number: 134-15081)
- Urea (Wako Pure Chemicals, catalog number: 219-00175)
- Silver Stain Kit (Wako Pure Chemicals, catalog number: 291-50301, compatible with MS)
- Trifluoroacetic acid (TFA)
- NADH (Oriental Yeast, catalog number: 44320000)
- Labeling buffer (see Recipes)
- 4x BN-PAGE sample buffer (see Recipes)
- Elution buffer (see Recipes)
- 4x Schägger sample buffer (see Recipes)
- 10x cathode buffer (see Recipes)
- 10x anode buffer (see Recipes)
- AB-3 mix (see Recipes)
- 3x gel buffer (see Recipes)
- 1.0 M KPi buffer, pH 7.4 (see Recipes)
- Gel composition for dSDS-PAGE (see Recipes)
Equipment
- Pippetmann (Gilson)
- -80 °C freezer (PHC, model: MDF-DU300H)
- HPLC system (Shimazdu, model: LC-10AS)
- Micro-centrifuge (CHIBITAN, Merck Millipore, model: XX42CF0RT)
- γ-counter (COBRATM II, Packard)
- Vacuum centrifugal evaporator (EYELA, model: CVE-2200)
- Bio-imaging analyzer (FLA-5100 or Typhoon FLA-9000 , Fuji Film or GE healthcare, respectively)
- Imaging plate (BAS IP MS 2040E, GE Healthcare)
- Vortex mixer (AS ONE, model: Trio TM-1N)
- Heat block (AS ONE, model: MyBL-10)
- Long wavelength UV lamp (UVP, Black-lay model B-100AP, equipped with a 100 W bulb, 365 nm)
- Electro-Eluter (Bio-Rad, model: 422)
- Gel dryer (Bio-Rad, Model583, equipped with HydroTech vacuum pump)
- Gel electrophoresis apparatus and glass plates (ATTO, AE-6500, 8 x 9 cm for mini-size gel format)
Software
- Image analysis software such as Multi Gauge or Image Quant (Fuji Film or GE Healthcare, respectively)
Procedure
文章信息
版权信息
© 2019 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Murai, M. and Miyoshi, H. (2019). Photoaffinity Labeling of Respiratory Complex I in Bovine Heart Submitochondrial Particles by Photoreactive [125I] amilorides. Bio-protocol 9(17): e3349. DOI: 10.21769/BioProtoc.3349.
- Uno, S., Kimura, H., Murai, M. and Miyoshi, H. (2019). Exploring the quinone/inhibitor-binding pocket in mitochondrial respiratory complex I by chemical biology approaches. J Biol Chem 294(2): 679-696.
分类
生物化学 > 其它化合物 > 小分子药物
生物化学 > 蛋白质 > 标记
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link