- EN - English
- CN - 中文
Assessment of Metacaspase Activity in Phytoplankton
浮游植物中半胱天冬酶活性检测
发布: 2019年08月20日第9卷第16期 DOI: 10.21769/BioProtoc.3341 浏览次数: 5111
评审: Alba BlesaJörg ToepelDavid A. Cisneros
Abstract
Programmed cell death (PCD) is an irreversible, genetically-controlled form of cell suicide in which an endogenous biochemical pathway leads to morphological changes and ultimately, cellular demise. PCD is accompanied by de-novo protein synthesis of a family of proteases-"caspases" that are often used as a diagnostic marker of PCD. Although phytoplankton do not contain true caspases, caspase-like activity (hypothetical proteins with analogous activity) has been traditionally used as a diagnostic marker of PCD in marine phytoplankton. Increased caspase-like proteolytic activity was demonstrated when synthetic fluorogenic activity substrates specific for caspases (with an Asp at the P1 position) were applied upon PCD induction. Metacaspases, cysteine proteases, share structural properties with those of caspases, yet they are highly specific for Arg and Lys cleavage site at the P1 position implying that caspase specific substrates are not indicative of metacaspase catalytic activity. This method specifically tests direct metacaspase activity in phytoplankton by the cleavage of the fluorogenic metacaspase substrate Ac-VRPR-AMC. Metacaspase activity was tested by the addition of a metacaspase specific peptide that is conjugated to the fluorescent reporter molecule. The cleavage of the peptide by the metacaspase releases the fluorochrome that, when excited by light, emits fluorescence. The level of metacaspase enzymatic activity in the cell lysate is directly proportional to the fluorescence signal detected. The use of specific standards in this test enables the quantification of the fluorescence results. This assay directly allows monitoring the metacaspase cleavage products and thereby tracing evidence for programmed cell death.
Keywords: Metacaspase activity (半胱天冬酶活性)Background
PCD is an irreversible, genetically-controlled form of cell suicide which is a fundamental feature of prokaryotic and eukaryotic microbial life essential for the regulation of cellular and tissue homeostasis in metazoans (Aravind et al., 1999). In phytoplankton, PCD has been defined as a form of autocatalytic cell suicide in which an endogenous biochemical pathway leads to morphological changes and ultimately, cellular dissolution (Bidle and Falkowski, 2004). PCD has been demonstrated in diverse evolutionary lineages of prokaryotic and eukaryotic phytoplankton including cyanobacteria, coccolithophores, diatoms, and dinoflagellates (Vardi et al., 1999; Segovia et al., 2003; Berman-Frank et al., 2004; Bidle and Falkowski, 2004; Bidle and Bender, 2008). In phytoplankton, environmental stresses, such as cell age, nutrient stress, excessive salt concentrations or oxidative stress and viral attacks may initiate PCD (Berman-Frank et al., 2004; Bidle and Falkowski, 2004; Spungin et al., 2019). This process is accompanied by de novo protein synthesis of a family of proteases-the caspases, (cysteine aspartic proteases) cleaving specifically at Asp residues at the P1 position. Caspases are often used as a diagnostic marker of PCD. Classic caspases are unique in metazoans and in several viruses (Minina et al., 2017). Higher plants, unicellular protists, fungi, and bacteria lack true caspases but contain metacaspases (Uren et al., 2000), cysteine-proteases that share structural properties, specifically a histidine-cysteine catalytic dyad in the predicted active site (Tsiatsiani et al., 2011). Metacaspases are widespread among prokaryotic and eukaryotic phytoplankton genomes. It has been indicated that prokaryotic phytoplankton, as well as evolving eukaryotic lineages, not only express PCD that involves caspase-like activity, but also a core set of proteins that are orthologues of metazoan caspases (Bidle and Falkowski, 2004; Bidle, 2016; Koonin and Aravind 2002).
Although phytoplankton do not contain true caspases, upon PCD induction caspase proteolytic activity is demonstrated when synthetic fluorogenic activity substrates are applied with an Asp at the P1 position (Berman-Frank et al., 2004 and 2007; Bidle and Bender, 2008; Thamatrakoln et al., 2012; Bar-Zeev et al., 2013). Unlike caspases, metacaspases lack Asp specificity and cleave their peptide substrates after Arg or Lys in the P1 position (Tsiatsiani et al., 2011). This implies that caspase specific substrates are not indicative of metacaspase catalytic activity. Thus, it has been suggested to use substrates with Arg or Lys residues at the P1 position to detect metacaspase activities in cellular extracts (Tsiatsiani et al., 2011; Klemenčič et al., 2015). Recently developed substrates applied to investigate the activity of specific metacaspases (Tsiatsiani et al., 2011; Klemenčič et al., 2015) have enabled further examination into the roles of metacaspases in organisms possessing metacaspase genes.
The method we detail herewith specifically tests direct metacaspase activity in phytoplankton by examining the cleavage of the fluorogenic metacaspase substrate Ac-VRPR-AMC (Ac-Val-Arg-Pro-Arg-AMC). Metacaspase activity tests and method calibrations were performed specifically in the marine cyanobacteria Trichodesmium under conditions that induce PCD (e.g., iron starvation and oxidative stress) and in samples collected from the South West Pacific Ocean (Spungin et al., 2018; Spungin et al., 2019). In this method, Trichodesmium cell lysate was tested for protease activity by the addition of a metacaspase specific peptide that is conjugated to the fluorescent reporter molecule, for example, 7-amino-4-methyl coumarin (AMC). The cleavage of the peptide by the metacaspase releases the fluorochrome that, when excited by light at 380 nm wavelength, emits fluorescence at 460 nm. Enzymatic activity in the cell lysate is directly proportional to the fluorescence signal detected with a fluorimeter or a fluorescent microplate reader.
Materials and Reagents
- Pipette tips
- Polycarbonate filters (e.g., PC 25 mm 5 µm, Whatman 110613)
- Microcentrifuge tubes (e.g., Axygen, catalog number: 3110451)
- Ice
- 96-well plate-black (e.g., SPL, Cell Culture Plate, PS, 96-well, Black, catalog number: 30496)
- HEPES (Sigma-Aldrich, catalog number: H3375)
- Sodium chloride (e.g., Merck, catalog number: 105404)
- Sucrose (e.g., AMRESCO, catalog number: 0335)
- CHAPS (3-(3-cholamidopropyl)-dimethylammonio-1-propanesulfonate) (Sigma-Aldrich, catalog number: C9426)
- Dithiothreitol (DTT) (Bio-Rad, catalog number: 1610611)
- Dimethyl Sulfoxide (DMSO) (Sigma-Aldrich, catalog number: 154938)
- Fluorogenic metacaspase substrate, Ac-VRPR-AMC, Mw = 725.85
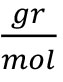 (BACHEM, catalog number: 4048494), store at -20 °C
(BACHEM, catalog number: 4048494), store at -20 °C - 7-amino-4-methyl coumarin (AMC) standard (Sigma-Aldrich, catalog number: A9891), Mw = 175.18
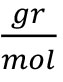 , store at -20 °C
, store at -20 °C - PierceTM BCA protein assay kit (Thermo, catalog number: 23225)
- Liquid nitrogen
- 5x Lauber buffer (see Recipe 1)
- 1 M AMC standard stock (see Recipe 2)
- 3 M Fluorogenic metacaspase substrate, Ac-VRPR-AMC (see Recipe 3)
- Working solution (Fluorogenic metacaspase aubstrate) (see Recipe 4)
Equipment
- Pipettes
- Ice bucket
- Forceps
- -20 °C freezer
- -80 °C freezer
- Ultra-cell disruptor (e.g., Sonic Dismembrator, Fisher Scientific, Waltham, MA)
- Microcentrifuge (e.g., Thermo Heraeus Pico 17 Microcentrifuge, Thermo Fisher, catalog number: 75002410)
- Vortex (e.g., Labnet, model: VX100 Vortex Mixer, catalog number: S-0100)
- Microplate reader equipped with fluorescence detection capabilities, kinetic reads and shake options (e.g., Synergy4 BioTek, Winooski, VT)
Procedure
文章信息
版权信息
© 2019 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Spungin, D. and Berman-Frank, I. (2019). Assessment of Metacaspase Activity in Phytoplankton. Bio-protocol 9(16): e3341. DOI: 10.21769/BioProtoc.3341.
分类
微生物学 > 微生物生物化学 > 蛋白质 > 活性
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link