- EN - English
- CN - 中文
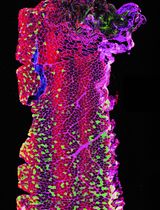
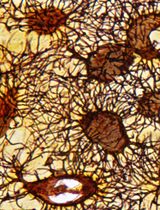
Looking through Brains with Fast Passive CLARITY: Zebrafish, Rodents, Non-human Primates and Humans
利用 Fast Passive CLARITY技术研究斑马鱼、啮齿动物、灵长类动物和人类的大脑
发布: 2019年08月05日第9卷第15期 DOI: 10.21769/BioProtoc.3321 浏览次数: 9509
评审: Oneil G. BhalalaBin ZhangAnonymous reviewer(s)
Abstract
Recently developed CLARITY (Clear Lipid-exchanged Acrylamide-hybridized Rigid Imaging/Immunostaining/In situ-hybridization-compatible Tis-sue-hYdrogel) technique renders the tissue transparent by removing lipids in the tissue, while preserving and stabilizing the cellular and subcellular structures. This provides effective penetration of diverse labeling probes, from primary and secondary antibodies to complementary DNA and RNA strands. Followed by high-resolution 3D imaging of neuronal cells and their projections in thick sections, tissue blocks, whole brains, or whole animals, CLARITY allows for superior quantitative analysis of neuronal tissue. Here, we provide our detailed protocol for PACT (Passive Clarity Technique) in brain tissue of diverse species, including human, non-human primate, rodents, and zebrafish. We describe the six principal steps: (1) Tissue fixation and preparation, (2) Passive lipid removal, (3) Immuno-labeling, (4) Optical clearing, (5) Imaging, (6) 3D visualization and quantification.
Keywords: CLARITY (CLARITY)Background
Optimization of tissue imaging techniques had to overcome several inherent problems, including lack of optical tissue transparency and spatial limits on antibody penetration. The solutions to these problems have evolved over time. Major advances in microscopy provided for superior 2D and 3D image resolution (Richardson and Lichtman, 2015; Whitehead et al., 2017). Thin sections, less than 40 microns in thickness, allowed most of the antibodies to reach their target proteins. Combined with diverse fluorescent tags, these antibodies revealed tissue complexities beyond those known before. However, evaluation of a tissue block or whole organ divided into many thin sections remained far from trivial. Following the time-consuming process of cutting and staining individual sections, an inherent discontinuity of specimens required the development of sophisticated imaging reconstruction techniques for accurate quantification of cells and ability to follow their individual projections, e.g., micro-optical sectioning tomography (MOST) (Li et al., 2010). Finally, to increase tissue transparency, a number of strategies to “clear” the tissue has been proposed, first by Werner Spalteholz as early as 1914 (Spalteholz, 1914) and by many other researchers thereafter (Efimova and Anokhin, 2009; Hama et al., 2011; Ertürk et al., 2012; Ke et al., 2013; Susaki et al., 2014; Fumoto et al., 2016). Despite the effectiveness of these strategies, there were limitations in tissue types and species in which they performed best, with some causing tissue shrinking (for review, Mano et al., 2018). Other technical difficulties include the rate and volume at which antibodies penetrated into a thick cleared tissue block or section, or whole organ specimen for labeling of proteins, and adverse effects of time and/or exposure to light on fluorescence emission.
In 2013, Stanford researchers Kwanghun Chung and Karl Deisseroth developed a novel approach called CLARITY (Clear Lipid-exchanged Acrylamide-hybridised Rigid Imaging/Immunostaining/In situ-hybridization-compatible Tis-sue-hYdrogel) (Chung et al., 2013; Chung and Deisseroth, 2013). By simultaneously removing lipids and infusing the entire protein structure with a hydrogel, CLARITY preserved the tissue architecture, proteins and nucleic acid molecules, while making a large tissue block or an entire organ optically transparent. Importantly, the removal of lipids using this method enhanced antibody penetration into the preserved tissue, facilitating immunohistochemical staining, allowing for more efficient and accurate quantitative analysis. The success of CLARITY is highlighted by its increasing popularity among neuroscientists and biologists studying diverse tissues and organs (Azaripour et al., 2016; Mortazavi et al., 2016; Jensen and Berg, 2017; Vigouroux et al., 2017; Du et al., 2018; Yu et al., 2018).
Active use of CLARITY technique resulted its further optimization, including PACT (passive CLARITY technique) and PARS (perfusion assisted agent released in situ), or ACT-PRESTO (active cleaning technique pressure related efficient and stable transfer of macromolecules into organs) (Yang et al., 2014; Tomer et al., 2014; Lee et al., 2016). These methods proved to be applicable to a diverse array of tissues, including the peripheral organs such as the liver, kidney, intestine and lung (Lee et al., 2014; Font-Burgada et al., 2015; Neckel et al., 2016; Saboor et al., 2016). While there are common features in CLARITY methodology, the processing and imaging of diverse tissues, organs or whole animals may differ between model organisms. Human tissues also require special considerations due to the high lipid content of human brain tissue, and often the prolonged post-mortem interval (PMI) that can affect the quality of tissue, and its fixation.
Here we share our protocols for using CLARITY to visualize a number of proteins of interest in brain tissue of several species, including zebrafish, rat, mouse, rhesus monkey, and human. We find the technique to be relatively simple to execute, highly efficient in clarifying whole zebrafish, individual brains, large brain tissue blocks or thick sections. We also find that our CLARITY protocol allows for using lower than earlier reported antibody concentrations to effectively reveal target proteins, enabling high-quality 3D visualization. In addition to earlier proposed semi-quantitative analysis of CLARITY-processed whole-brain zebrafish samples, based on fluorescence Intensity (Lindsey and Kaslin, 2017), we show that 3D analytical tools (e.g., Fiji or Imaris) can provide accurate counts and morphological parameters of labeled cells, axons, dendrites, or any other quantitative immunohistochemical labeling. Together, we find CLARITY to be an exceptional tool for 3D visualization and quantification of brain tissue constituents, which can further be used in studies of neurogenesis, connectivity, and pathological brain conditions.
Materials and Reagents
- Tissue Specimens tested in this Protocol
- Zebrafish (Danio rerio): whole animal, brain (compatible with any age)
- Rat (Rattus or Mus musculus): brain (compatible with any age)
- Non-Human Primate (e.g., Macaca mulatta): brain (compatible with any age)
- Human: brain (anatomical donation, compatible with any age)
- CLARITY Supplies and Reagents
- 1.5 ml Eppendorfs (USA Scientific, catalog number: 1615-5500)
- 50 ml FalconTM Tubes (Fisher Scientific, catalog number: 14-432-22)
- DWK Life Sciences KimbleTM 7 ml Solvent-SaverTM Scintillation Vials (Fisher Scientific, catalog number: 03-340-128)
- Thermo ScientificTM NalgeneTM Rapid-FlowTM Sterile Disposable Bottle Top Filters with PES Membrane, 0.45 µm (Fisher Scientific, catalog number: 09-740-64A)
- Razorblades (Fisher Scientific, catalog number: 12-640)
- Phosphate Buffered Saline (PBS) 10x, Fisher BioReagents, pH 7.4 (Fisher Scientific, catalog number: BP3994) (store at room temperature)
- Sodium hydroxide solution (1 N) (NaOH) (Fisher Scientific, catalog number: SS2661) (store at room temperature)
- Formalin solution, neutral buffered 10% (Sigma-Aldrich, catalog number: HT501128-4L) (store at room temperature)
- 200 mg/L MS222 (see Recipes)
Tricaine (MS222) (Sigma-Aldrich, catalog number: E10521-10G) (store at room temperature) - 0.1 M PBS with 0.02% sodium azide (pH 7.4) (see Recipes)
Sodium Azide (Sigma-Aldrich, catalog number: S2002) (store at room temperature) - Paraformaldehyde (PFA) Fixing solution (4% PFA, 1x PBS,) pH 7.4 (see Recipes)
Paraformaldehyde (PFA) (Sigma-Aldrich, catalog number: 252549) (store at room temperature) - CLARITY Solution (pH 8.5) (see Recipes)
- Boric Acid (Sigma-Aldrich, catalog number: B7901)
- Sodium dodecyl sulfate (SDS) (Sigma-Aldrich, catalog number: L3771-500G)
- Lithium hydroxide monohydrate (Sigma-Aldrich, catalog number: 254274)
- 0.05 M TBS (pH 8.0) (see Recipes)
Tris Buffer Saline (TBS) (Sigma-Aldrich, catalog number: T6664) (store at room temperature) - 2 M Hydrochloric acid (HCI) (see Recipes)
Hydrochloric acid, 37% for analysis (HCI) (Acros Organics, catalog number: 450550025) (store at room temperature) - 0.1 M boric acid (pH 8.5) (see Recipes)
- Immunohistochemistry Reagents
- Click-iTTM EdU Alexa FluorTM 488 Flow Cytometry Assay Kit (Thermo Fisher Scientific, catalog number: C10420)
- CuSO4 (store at 4 °C)
- Fluorescent dye azide (store at -20 °C)
- Reaction Buffer Additive (store at -20 °C)
- 0.05 M TBS (pH 8.0) with 1% Triton X-100 (see Recipes)
Triton X-100 (Sigma-Aldrich, catalog number: X100) (store at room temperature) - Antibodies (Tables 1 and 2)
Table 1. Primary antibodies used in this Protocol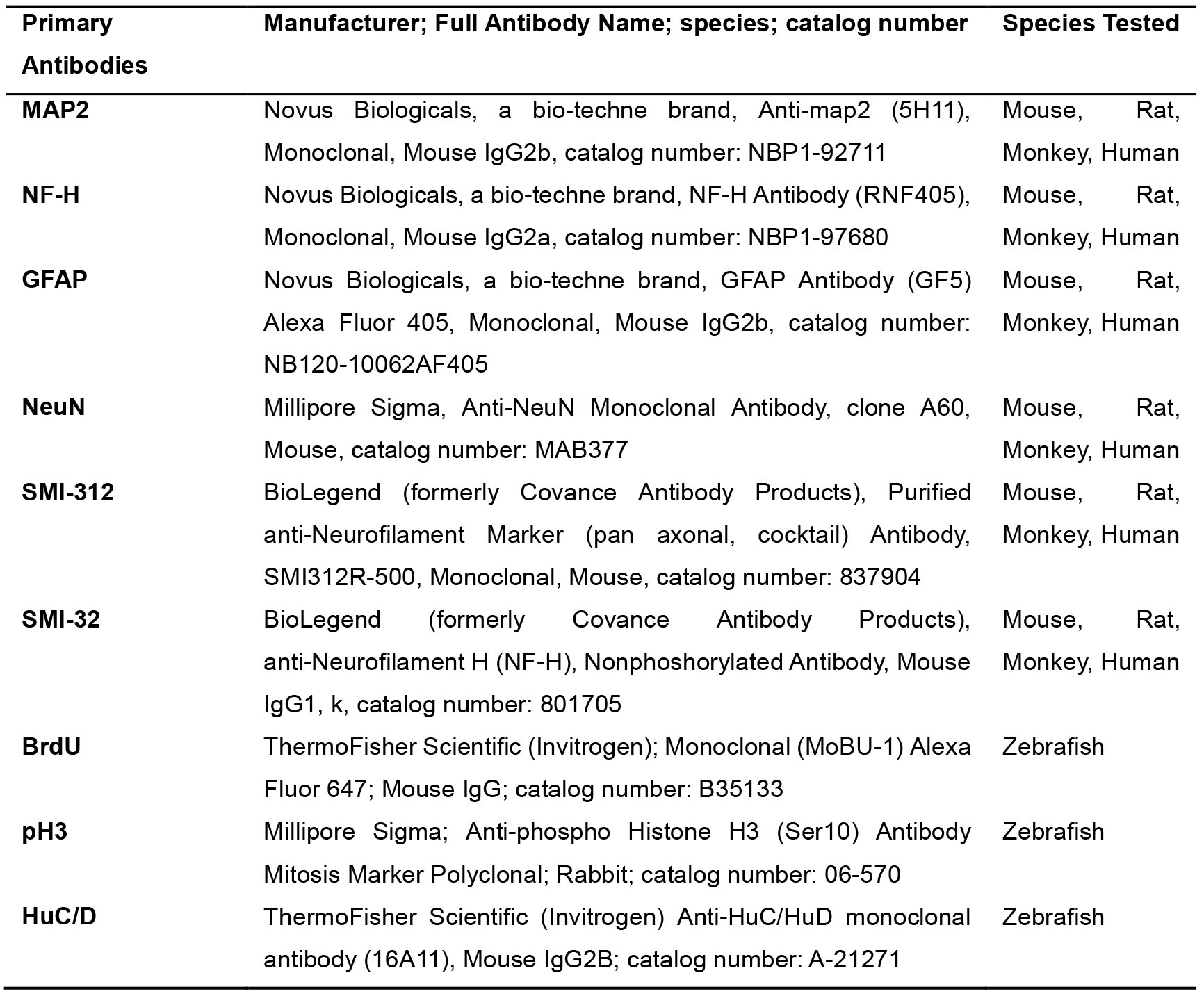
Table 2. Secondary antibodies used with this Protocol
- Sodium citrate buffer (store at room temperature, see Recipes)
Tri-sodium citrate (dihydrate) (Fisher Scientific, catalog number: 78-101-KG) - 50% formamide/50% 2x SSC (see Recipes)
a.Formamide Certified ACS, Fisher Chemical (Fisher Scientific, catalog number: 75-12-7) (store at 4 °C)
b.20x SSC Buffer (Ambion, catalog number: AM9763) (store at room temperature) - EdU labeling solution (see Recipes)
- Click-iTTM EdU Alexa FluorTM 488 Flow Cytometry Assay Kit (Thermo Fisher Scientific, catalog number: C10420)
- Imaging Supplies and Reagents
- Aluminum foil (Sigma-Aldrich, catalog number: 326852)
- Paint Brush
- 15 ml polypropylene conical tubes (Falcon, Fisher Scientific, catalog number: 352096)
- Blu Tack (pliable adhesive that can be readily formed into any shape, Amazon.com)
- Pelco Black Wall Glass Bottom Dishes (Ted Pella, catalog number: 14032-120)
- Camco 25573 Bullseye Level (Amazon)
- 80% glycerol solution in 0.1 M PBS (see Recipes)
Glycerol (Sigma-Aldrich, catalog number: G7893) (store at room temperature)
Equipment
- Microscopes:
- Upright Stereoscope Microscope (e.g., Zeiss)
- Confocal Microscope (e.g., Leica SP8 using an HC Fluotar 25x/0.96 W VISIR objective)
- Stirring Hot plate (e.g., Fisher Scientific, catalog number: 6796220)
- Chemical hood
- Vibratome (e.g., Pelco 102 Vibratome Sectioning System)
- Rocker (orbital or tube rocker)
- Incubator (37 °C)
- Forceps (e.g., Fisher Scientific, catalog number: 12000122)
Software
- ImageJ/Fiji (NIH, Bethesda, Maryland), Bitplane by Imaris, LasX (Leica Software)
Procedure
文章信息
版权信息
© 2019 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Mortazavi, F., Stankiewicz, A. J. and Zhdanova, I. V. (2019). Looking through Brains with Fast Passive CLARITY: Zebrafish, Rodents, Non-human Primates and Humans. Bio-protocol 9(15): e3321. DOI: 10.21769/BioProtoc.3321.
- Stankiewicz, A. J., Mortazavi, F., Kharchenko, P. V., McGowan, E. M., Kharchenko, V. and Zhdanova, I. V. (2019). Cell kinetics in the adult neurogenic niche and impact of diet-induced accelerated aging. J Neurosci 39(15): 2810-2822.
分类
神经科学 > 神经解剖学和神经环路 > 荧光成像
细胞生物学 > 组织分析 > 组织成像
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link