- EN - English
- CN - 中文
Axon-seq for in Depth Analysis of the RNA Content of Neuronal Processes
神经元突起RNA含量的Axon-seq 深入分析
发布: 2019年07月20日第9卷第14期 DOI: 10.21769/BioProtoc.3312 浏览次数: 12404
评审: Gal HaimovichShyam SolankiAnonymous reviewer(s)
Abstract
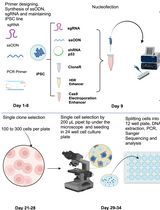
Neuronal processes have an RNA composition that is distinct from the cell body. Therefore, to fully understand neuronal biology in health and disease we need to study both somas, dendrites and axons. Here we describe a detailed protocol of a newly refined method, Axon-seq, for RNA sequencing of axons (and dendrites) grown in isolation using single microfluidic devices. We also detail how to generate motor neurons from mouse and human pluripotent stem cells for sequencing, but Axon-seq is applicable to any neuronal cell. In Axon-seq, the axons are recruited through a growth factor gradient, lysed and directly processed to cDNA without RNA isolation. A careful bioinformatic step ensures that any soma-contaminated samples are easily identified and removed.
Keywords: RNA sequencing (RNA测序)Background
Neurons are highly polarized cells. Their processes, both dendrites and axons, need to be able to respond to changes in the microenvironment in a manner independent of the soma (Holt and Schuman, 2013). To fully understand neuron biology, it is therefore important to be able to study neurites in isolation, in addition to conducting analysis of the cell bodies. This appears particularly important for neurons where the axon and dendrites constitute the majority of the cellular volume, as is the case for spinal motor neurons where we estimate that they comprise approximately 99% of the cellular volume. To isolate neurites, there are excellent tools such as Campenot Chambers (Boyden, 1962; Campenot, 1977) or microfluidic chambers (Taylor et al., 2005).
However, while neurites are separated out in such devices, it is important to bear in mind that cross-contamination between compartments can still occur. Thus, RNA sequencing of isolated axonal compartments (Minis et al., 2014; Saal et al., 2014; Briese et al., 2016; Rotem et al., 2017) can lead to incorrect results/conclusions when the purity and exclusion of somas is insufficiently examined. To ensure detailed and accurate investigation of motor axonal mRNA composition and its modulation in ALS we developed Axon-seq (Nijssen et al., 2018). This is an application of our LCM-seq method for single-cell spatial RNA-sequencing (Nichterwitz et al., 2016).
In Axon-seq, we use microfluidic devices to separate axons from stem cell-derived motor neurons (mouse and human) from their somas. As motor neurons have axons that traverse far longer distances than their dendrites, we are able to analyze axons alone. This may not be possible for all neuronal subtypes, but the method still allows for analysis of neurites as an entity.
In contrast to previous methods, Axon-seq does not require an RNA isolation step, and it allows for high sensitivity and cost-efficient sequencing from a single microfluidic device.
Notably, Axon-seq effectively eliminates all samples with any somatic cross-contamination, as it uses a highly stringent and sensitive bioinformatic quality control step that identifies axonal samples containing trace levels of mRNA from undesired cell somas. Here we provide a detailed protocol for Axon-seq which can be applied to any cell containing longer processes.
Materials and Reagents
- Materials
- Sterile 6 and 10 cm bacterial dishes (non-adhesive plastic) (Corning, catalog numbers: 351007 and 351029)
- Pipette tips (0.5-10 μl, 10-100 μl, 100-1,000 μl)
- Sterile 15 and 50 ml Falcon tubes
- Parafilm
- 70 μm cell-strainer filter (VWR, catalog number: 732-2758P)
- 24 x 32 mm cover glasses (VWR, Menzel Gläser, catalog number: 630-2873)
- Soft pencil-type brush (Any soft brush type that does not have too strong hairs that could pierce/damage the PDMS-surface of which the microfluidic devices are made. Brushes, from a typical hardware/convenience store, with a diameter of 2-5 mm work best, laboratory grade is not required. Nylon/synthetic hairs are not an issue.)
- Waterproof marker with fine (1 mm) or very fine (0.3 mm) tip
- Biological material
- Motor neurons from mouse embryonic stem cells
- Motor neurons from human pluripotent stem cells
- Reagents
- 70% ethanol
- Triton X-100 (Sigma-Aldrich, catalog number: X100)
- PBS (Thermo Fisher, catalog number: 14190136)
- Poly-L-ornithine, 0.01% solution (Sigma-Aldrich, catalog number: A-004-C)
- Fibronectin, 1 mg/ml solution (Sigma-Aldrich, catalog number: F1056)
- Laminin, 1 mg/ml solution (Sigma-Aldrich, catalog number: L2020)
- TrypLe Express (Thermo Fisher, catalog number: 12604021)
- Neurobasal (Thermo Fisher, catalog number: 21103049)
- DMEM/F12 with GlutaMAX (Thermo Fisher, catalog number: 31331028)
- B27 supplement, custom (Thermo Fisher, catalog number: 0080085SA)
- Penicillin/Streptomycin (Thermo Fisher, catalog number: 15140122)
- GlutaMAX supplement 100x (Thermo Fisher, catalog number: 35050061)
- 2-Mercaptoethanol (Sigma-Aldrich, catalog number: M6250)
- Ascorbic acid, 100 mg powder (Sigma-Aldrich, catalog number: A4403), prepare as 200 mM stock solution by dissolving the powder in 2.84 ml of nuclease-free dH2O
- Y-27632, 10 mg powder (Tocris, catalog number: 1254), prepare as 10 mM stock solution by dissolving the powder in 3.12 ml nuclease-free dH2O
- LDN-193189 dihydrochloride, 10 mg powder (Tocris, catalog number: 6053), prepare as 10 mM stock solution by dissolving the powder in 2.09 ml DMSO, then prepare 1 mM working stocks (1:10 diluted) in DMSO
- SB-431542, 10 mg powder (Tocris, catalog number: 1614), prepare as 10 mM stock solution by dissolving the powder in 2.60 ml DMSO
- CHIR-99021, 10 mg powder (Tocris, catalog number: 4423), prepare as 3 mM stock solution by dissolving the powder in 7.16 ml DMSO
- DAPT, 10 mg powder (Tocris, catalog number: 2634), prepare as 10 mM stock solution by dissolving the powder in 2.31 ml DMSO
- SAG, 1 mg powder (Tocris, catalog number: 4366), prepare as 500 μM stock solution by dissolving the powder in 4.08 ml DMSO
- All-trans retinoic acid, 50 mg powder (Sigma-Aldrich, catalog number: R2625), prepare as 100 mM stock solution by dissolving the powder in 1.66 ml DMSO, then prepare 1 mM working stocks (1:100 diluted) in DMSO
- Dimethyl sulfoxide (DMSO) (Sigma-Aldrich, catalog number: D2650)
- Bovine serum albumin (BSA), 7.5% solution in dPBS (Sigma-Aldrich, catalog number: A8412)
- Glial-derived neurotrophic factor (GDNF), 10 μg powder (Peprotech, catalog number: 450-10), prepare as 10 μg/ml stock solution by dissolving the powder in 1 ml dPBS with 0.1% BSA
- Brain-derived neurotrophic factor (BDNF), 10 μg powder (Peprotech, catalog number: 450-02), prepare as 10 μg/ml stock solution by dissolving the powder in 1 ml dPBS with 0.1% BSA
- RNase-inhibitor (Takara, catalog number: 2313A)
- DTT (part of SuperScript II Reverse Transcriptase kit, Thermo Fisher, catalog number: 18064071)
- Nuclease-free dH2O (Thermo Fisher, catalog number: 10977-035)
- 37% formaldehyde solution (Merck Millipore, catalog number: 818708)
- Mouse MN medium (see Recipes)
- N2 medium (see Recipes)
- B27 medium (see Recipes)
- Human EB medium (see Recipes)
- Lysis/harvesting solution (see Recipes)
Equipment
- Glass beaker (500 ml)
- Sterile forceps (Dumont, catalog number: 11251-20, or other forceps of similar standard size)
- Pipettes (0.5-10 μl, 10-100 μl, 100-1,000 μl)
- Magnetic stirrer
- Sterile laminar flow hood
- Humidified incubator (37 °C, 5% CO2)
- Suction aspirator
- Standard Neuron Device (Xona MicrofluidicsTM, SND75/150/450/900)
- Low-speed orbital shaker (Elmi-tech, DOS-20S)
- Inverted bright field microscope (> 20x magnification is required to reliably see axons exiting the microgrooves. Examples are Laxco LMI-3000 or Carl Zeiss PrimoVert. Any similar setup is fine.)
Software
- R (https://www.r-project.org)
- RStudio open source edition, Boston, MA, USA (https://www.rstudio.com)
Procedure
文章信息
版权信息
© 2019 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Nijssen, J., Aguila, J. and Hedlund, E. (2019). Axon-seq for in Depth Analysis of the RNA Content of Neuronal Processes. Bio-protocol 9(14): e3312. DOI: 10.21769/BioProtoc.3312.
分类
神经科学 > 细胞机理 > RNA 定位
干细胞 > 多能干细胞 > 基于细胞的分析方法
分子生物学 > RNA > RNA 测序
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link