- EN - English
- CN - 中文
Total RNA Isolation from Separately Established Monolayer and Hydrogel Cultures of Human Glioblastoma Cell Line
单层水凝胶培养人胶质母细胞瘤细胞系的全RNA分离
发布: 2019年07月20日第9卷第14期 DOI: 10.21769/BioProtoc.3305 浏览次数: 6218
评审: Oneil G. BhalalaSébastien GillotinAnonymous reviewer(s)
Abstract
Astrocytoma is an invasive carcinoma occurring in the nervous system and currently lacks effective treatment options. A deeper understanding of the mechanisms of tumorigenesis and tumor progression is needed in order to develop novel therapeutic strategies. Recent advances in in vitro culture systems have demonstrated that the use of three-dimensional (3D) culture models could be more relevant for this purpose as compared to monolayer or two-dimensional (2D) models due to their resemblance to in vivo cancer pathology. High-throughput techniques such as RNA sequencing, microarray analyses and cloning could provide useful insights into the relevance of these systems to the native tissue. Previous studies have reported RNA extraction protocols needed for such applications. We have modified these protocols to suit the isolation of total RNA from monolayer and hydrogel cultures of astrocytoma established using basement membrane matrix, GeltrexTM. We have used this method to demonstrate the differences in the expression of genes involved in autophagy, a process deregulated in many cancer types, in monolayer and hydrogel cultures using quantitative polymerase chain reaction (qPCR). This protocol can be adopted by the researchers who wish to understand the molecular basis of gene expression in hydrogel cultures of normal as well as cancer cell lines.
Keywords: RNA (核酸)Background
Astrocytomas are central nervous system tumors of glial origin. Poor survival rates among patients within 5 years of diagnosis and almost inevitable relapse are the hallmarks of this life-threatening disease (Krex et al., 2007). Our understanding of disease pathogenesis is limited, resulting in the lack of effective intervention strategies. In vitro culture systems could prove useful for studying cell-cell interactions and intracellular signaling events in astrocytoma, and provide platforms for testing potential therapeutic compounds (Reardon et al., 2006; Baker and Chen, 2012). Three-dimensional (3D) in vitro culture platforms have especially gained popularity in recent years due to their biological relevance to in vivo tumors (Li et al., 2007; Whiteside, 2008; Tibbitt and Anseth, 2009; Xu et al., 2014; Jogalekar and Serrano, 2018). 3D cultures established using various tissue engineering scaffolds (e.g., hydrogels) allow for multi-layered growth of cells in a fashion similar to that of native tissue, unlike two-dimensional (2D) cultures, making them better predictors of responses to drug candidates.
Hydrogel culture systems could be exploited in parallel with monolayer cultures to further our understanding of cell-cell and cell-matrix interactions with high-throughput applications that analyze cellular gene expression, such as RNA-sequencing, microarray analysis and cloning. To accomplish this, a pure, intact, high quality RNA needs to be isolated in parallel from separately cultured monolayer and hydrogel cultures so that both cultures are processed under identical conditions and are therefore comparable. Several protocols have been reported for extracting RNA from 3D cultures established using a variety of commonly used tissue engineering scaffolds such as Matrigel and collagen (Lee et al., 2007; Mroue and Bissell, 2013). However, there is a paucity of information available with respect to RNA extraction from 3D cultures maintained in GeltrexTM, a relatively new basement membrane matrix derived from murine Engelbreth-Holm-Swarm tumors and manufactured by Invitrogen.
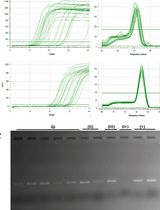
When we used existing protocols to carry out RNA isolation from multiple cell lines including astrocytoma, we found that our astrocytoma cell line (CCF-STTG1) was highly sensitive to the reagents that dissolve extracellular matrices used in 3D cultures and could not yield an intact RNA at the end of the procedure, although this procedure worked well with other mammalian and amphibian cell lines we used in our experiment. Therefore, we modified RNA isolation protocols available in the literature to extract total RNA from monolayer and hydrogel cultures of astrocytoma. We used this RNA to carry out gene expression analyses for key autophagy pathway genes, Beclin-1 (BECN1) and microtubule associated protein 1 light chain 3 beta (MAP1LC3B), using quantitative polymerase chain reaction (qPCR) (Jogalekar et al., 2017). Our findings indicate a modest upregulation of these genes in hydrogel cultures of astrocytoma and are consistent with previous reports suggesting their upregulation in correlation with poor prognosis in glioma patients (Pirtoli et al., 2009; Giatromanolaki et al., 2014).
Materials and Reagents
- 0.8 μm 150 ml filter unit (Thermo Scientific Nalgene, catalog number: 125-0080)
- NuncTM Cell Culture Treated Flasks with Filter Caps (NuncTM, catalog number: 136196)
- Falcon® Cell Scrapers, Sterile (Corning®, catalog number: 353085)
- Falcon® 50 ml High Clarity PP Centrifuge Tubes (Corning®, catalog number: 352098)
- RNase-free Microfuge Tubes (1.5 ml) (InvitrogenTM, catalog number: AM12400)
- 5cc BD Luer-LokTM Disposable Syringe (BD, catalog number: 309603)
- 21 G 1¼ needle (BD, catalog number: 305166)
- 10 ml serological pipette, sterile (USA Scientific, catalog number: 1071-0810)
- FisherbrandTM SureOneTM Aerosol Barrier Pipette Tips (FisherbrandTM SureOneTM, catalog numbers: 02-707-404 [1,000 μl], 02-707-430 [200 μl], 02-707-432 [20 μl])
- CCF-STTG1 cell line (ATCC, catalog number: CRL-1718)
- RPMI-1640 Medium (ATCC, catalog number: 30-2001)
- Fetal Bovine Serum (FBS) (ATCC, catalog number: 30-2020)
- LDEV-Free Reduced Growth Factor Basement Membrane Matrix, GeltrexTM (Invitrogen, catalog number: A1413202)
- 70% Ethanol
- RNaseZapTM RNase Decontamination Solution (AmbionTM, catalog number: AM9780)
- TRIzolTM Plus RNA Purification Kit (AmbionTM, catalog number: 12183555)
- Cell Recovery Solution (Corning, catalog number: 354253)
- Phosphate buffered saline (PBS) (Sigma-Aldrich, catalog number: P3813)
- Chloroform (Sigma-Aldrich, catalog number: C2432)
- Nuclease-Free Water (not DEPC-Treated) (AmbionTM, catalog number: AM9937)
- DNA-free DNA Removal Kit (AmbionTM, catalog number: AM1906)
- RNA 6000 Nano Kit (Agilent, catalog number: 5067-1511)
- Ethyl alcohol, Pure (Sigma-Aldrich, catalog number: E7023)
- RPMI-1640 complete medium (see Recipes)
- Sterile 1x PBS (see Recipes)
Equipment
- GilsonTM PIPETMAN ClassicTM Pipettes (Gilson, catalog numbers: F123600 [P20], F123601 [P200], F123602 [P1000], F144802 [P10], F144801 [P2])
- 2100 bioanalyzer instrument (Agilent, catalog number: G2939BA)
- Drummond Pipet-Aid, Plain, 110V (Drummond, catalog number: DP-110)
- Biosafety cabinet (Nuaire, catalog number: NU-425-600)
- 4 °C centrifuge (Beckman Coulter, catalog number: AllegraTM 21R)
- Centrifuge rotors
- Water bath at 37 °C (Sheldon Manufacturing Inc., catalog number: 1211)
- AutoFlow Water Jacket CO2 Incubator (Nuaire, catalog number: NU-4750)
- -20 °C freezer
Software
- 2100 Expert (Agilent)
- Photoshop CS6 (Adobe)
Procedure
文章信息
版权信息
© 2019 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Jogalekar, M. P. and Serrano, E. E. (2019). Total RNA Isolation from Separately Established Monolayer and Hydrogel Cultures of Human Glioblastoma Cell Line. Bio-protocol 9(14): e3305. DOI: 10.21769/BioProtoc.3305.
分类
癌症生物学 > 增殖信号转导 > 遗传学
神经科学 > 神经系统疾病 > 脑肿瘤 > 多形性胶质母细胞瘤
分子生物学 > RNA > RNA 提取
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link