- EN - English
- CN - 中文
An Adjustable Protocol to Analyze Chemical Profiles of Non-sterile Rhizosphere Soil
一种可调整的用于分析研究非无菌根际土壤化学特性的方法
发布: 2019年05月20日第9卷第10期 DOI: 10.21769/BioProtoc.3245 浏览次数: 6558
评审: Joëlle SchläpferYang BaiManish Kumar Patel
Abstract
The analysis of chemical diversity in non-sterile rhizosphere soil has been a pressing methodological challenge for years. Rhizosphere-enriched chemicals (i.e., rhizochemicals) include root exudation chemicals, (microbial) breakdown products thereof, and de novo produced metabolites by rhizosphere-inhabiting microbes, all of which can play an important role in plant-soil interactions. The power and resolution of analytical methods and statistical analysis pipelines allow for better acquisition, separation and identification of rhizochemicals, thus providing unprecedented insight into the biochemistry underpinning plant-soil interactions. The current protocol describes a recently developed method to characterize rhizochemical profiles from plants, including crops, and is modular and customizable, allowing for application across a range of different plant-soil combinations. The protocol provides in-depth details about the experimental system for sample collection, data acquisition by liquid chromatography coupled to mass spectrometry, and analytical pipeline, which statistically selects for rhizochemicals by statistical comparison between metabolite profiles from plant-containing soil and plant-free soil. Moreover, the optional addition of chemical standards permits a semi-targeted approach, which improves the annotation of chemical signatures and identification of single rhizochemicals.
Keywords: Metabolomics (代谢组学)Background
Previous approaches to studying rhizosphere have often focused on sterile and hydroponic growth conditions (van Dam and Bouwmeester, 2016). These approaches limit our understanding of the multi-trophic nature of the rhizosphere, as they fail to provide information about (microbial) breakdown products of root exudation chemicals and de novo produced chemicals by rhizosphere-inhabiting microbes, even though these may drive plant-soil interactions in response to environmental change, such as pathogen attack or abiotic stress. The challenge arises from understanding and simplifying the complex, and often overwhelming, level of chemical diversity that originates from untargeted analyses of non-sterile soil (Figure 1). Nevertheless, untangling this diversity by identifying chemical networks, their origin and function, is critical for obtaining new insight into the ecology of a much under-explored area of plant and soil science. Although not without its caveats, recent advances in mass spectrometry (MS) technology and statistical-analytical techniques permit powerful exploratory methods to uncover the chemistry of natural soils (Pétriacq et al., 2017; Swenson et al., 2015 and 2018). Such techniques are becoming increasingly accessible.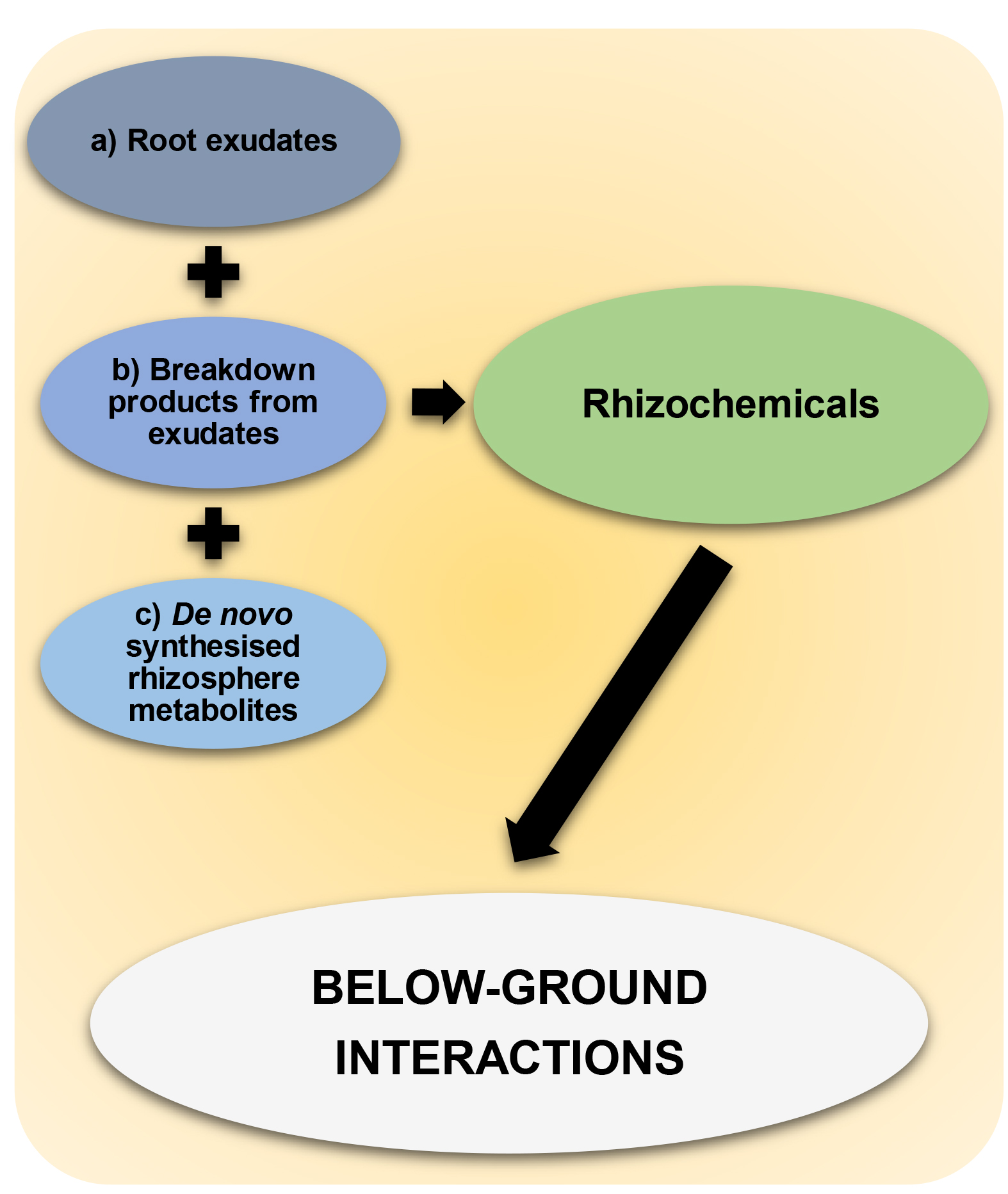
Figure 1. Rhizochemicals that shape below-ground interactions. Rhizochemicals are compounds that are enriched in the rhizosphere and include a) root exudation chemicals, b) microbial breakdown products of root exudation chemicals, c) de novo synthesized metabolites by rhizosphere-inhabiting microbes. Rhizochemicals can have important signaling activities thereby shaping below-ground interactions between plants and soil microbes.
The protocol presented here describes an adjustable experimental system for sample collection and sample preparation, followed by data acquisition using ultra-performance liquid chromatography coupled to quadrupole time-of-flight mass spectrometry (UPLC-Q-TOF). Subsequent statistical analysis allows separating chemical signatures by retention time and mass to charge (m/z) ratios between plant-free soil and plant-containing soil, which ultimately identifies profiles of rhizosphere-enriched chemical (i.e., rhizochemicals). These profiles can be further annotated using chemical databases, such as METLIN (Smith et al., 2005). Moreover, during mass spectrometry examination, running a library of relevant standards can increase the accuracy of this approach and allow focus of the analysis on specific metabolite classes.
The protocol outlined here, and originally used in Pétriacq et al. (2017), was initially developed for the analysis of rhizosphere chemistry in the model plant species Arabidopsis thaliana (Arabidopsis; Figures 2 and 3) and has been adapted for maize. For other plant species, a similar experimental design can be adapted and implemented, but method optimization is advised. Particularly, we recommend further optimization when insufficient separation is obtained between rhizochemical signatures from the plant-containing soil samples and the plant-free soil samples (see Figure 4). These adjustments involve i) testing different soils, ii) using different extraction solutions (e.g., with varying MeOH concentrations), iii) increasing root biomass in collection tubes, iv) adjusting soil moisture, v) increasing the time of sample extraction from the collection tubes, and vi) varying MS settings. Further attention should be paid to the fact that extraction solutions with high MeOH concentrations can cause damage to microbial and/or root cells, in which case specific assays can be used to assess the extent of cell damage, as detailed in Petriacq et al. (2017). If cell damage occurs, we recommend using a lower MeOH concentration in the extraction solution and/or shorter extraction times. If insufficient sample yield is obtained with shorter extraction times, a pressurized collection system can be used, as is illustrated in Figure 2D.
Materials and Reagents
Note: All chemicals and solvents used for metabolomics were of mass spectrometry grade (Sigma-Aldrich, Germany). Other solvents were of analytical grade.
- Tips
- Millipore miracloth, pore size of 22-25 µm (VWR, catalog number: 475855-1)
- Petri dish (lids) NunclonTM Delta, 8.8 cm2 (Thermo Fisher Scientific, catalog number: 150318)
- 15 ml FalconTM conical centrifuge tubes (Thermo Fisher Scientific, catalog number: 10468502)
- 50 ml FalconTM conical centrifuge tubes (Thermo Fisher Scientific, catalog number: 10788561)
- 50 ml syringe, without needle (Thermo Fisher Scientific, catalog number: 11901563)
- 5 ml conical preparation tube (Starlab, catalog number: E1450-1100)
- 2 ml microcentrifuge tube (Starlab, catalog number: I1420-2600)
- 30 ml SterilinTM universal container (Thermo Fisher Scientific, catalog number: 11309143)
- Aluminum foil
- Filter paper
- 0.2 µm disposable Merck cartridge (Thermo Fisher Scientific, catalog number: 10740365)
- 1.5 ml Glass screw neck vials, with screw caps and micro inserts (VWR, catalog numbers: 548-0018, -0020, -0106)
- Levington M3 dry compost (Spunhill, catalog number: YEV05K)
- Non staining silica sand (Sweepfast, catalog number: CH52)
- Arabidopsis seeds
- Maize seeds (Zea mays)
- Acetonitrile (Hypergrade, Sigma-Aldrich, catalog number: 1000291000)
- Methanol (Hypergrade, Sigma-Aldrich, catalog number: 1060351000)
- Formic acid (FA, Sigma-Aldrich, catalog number: F0507-500ML)
- Liquid Nitrogen (BOC)
- Bleach Mexcel (5-10%) (SLS, catalog number: CLE0300)
- Hydrochloric acid (37%) (Sigma-Aldrich, catalog number: H1758)
- Sodium formate (0.01 M NaOH/1% FA (1/1, v/v) ten-fold diluted with acetonitrile/water (80/20, v/v) (Sigma-Aldrich, catalog number: 71539-500G)
- Leucine enkephalin (2 ppm in acetonitrile/water (50/50, v/v) with 0.1% of formic acid) (Sigma-Aldrich, catalog number: L9133-25MG)
- Ice-cold extraction solution MeOH 50% + FA 0.05% (see Recipes)
Equipment
- Pipettes (P-5000, P-1000) (Starlab)
- Growth tubes (make these bespoke, see below)
- Pressure lids (make these bespoke, see below)
- Speedvac vacuum concentrator (Thermo Fisher Scientific, catalog number: SPD120)
- Freeze drier (Modulyo benchtop freeze dryer) (Edwards, catalog number: D-230)
- BEH C18 column for UPLC (2.1 x 50 mm, 1.7 μm, Waters) with a guard column (VanGuard, 2.1 x 5 mm, 1.7 µm, Waters)
- SYNAPT G2si Q-TOF (Waters)
- Centrifuge (that fits 5 ml tubes)
- Microcentrifuge
- Ultrasonic cleaning bath
- Soldering iron
- Drill (7 mm multipurpose drill bit)
- Growth cabinets (Fitotron, SANYO)
- Tweezers
- 4 °C refrigerator
- -80 °C freezer
Software
- MassLynx, version 4.1 with Data bridge, https://www.waters.com
- R with XCMS package, version 3.2, https://www.r-project.org
- Multiple experiment viewer–MeV, version 4.9.0, http://mev.tm4.org/#/welcome
- MetaboAnalyst version 3.0, http://www.metaboanalyst.ca
- METLIN, https://metlin.scripps.edu
Procedure
文章信息
版权信息
© 2019 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Williams, A., Ton, J. and Pétriacq, P. (2019). An Adjustable Protocol to Analyze Chemical Profiles of Non-sterile Rhizosphere Soil. Bio-protocol 9(10): e3245. DOI: 10.21769/BioProtoc.3245.
分类
植物科学 > 植物生物化学 > 其它化合物
植物科学 > 植物生理学 > 新陈代谢
生物化学 > 其它化合物 > 土壤中化学物质
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link