- EN - English
- CN - 中文
Quantification of Queuosine Modification Levels in tRNA from Human Cells Using APB Gel and Northern Blot
利用聚乙二醇凝胶和Northern印记法定量分析来自人细胞的tRNA 的Q核苷修饰水平
发布: 2019年03月20日第9卷第6期 DOI: 10.21769/BioProtoc.3191 浏览次数: 6838
评审: Gal HaimovichJoshua S TitlowAnonymous reviewer(s)
Abstract
Queuosine (Q) is a hypermodified base in the wobble anticodon position of tRNAs coding for the amino acids Tyr, His, Asn, and Asp. tRNA Q-modification is introduced by a queuine tRNA-ribosyltransferase (TGT) that replaces the guanine base at G34 at these tRNAs with the modified base. tRNA Q-modification is widely distributed among prokaryotic and eukaryotic organisms, but only bacteria synthesize Q-modified tRNA de novo. In mammals, tRNA Q-modifications strictly rely on the presence of gut microbiomes or diets to produce the queuine base. Despite decades of study, cellular roles of tRNA Q-modification are still not fully understood. Here we describe a method to quantify tRNA Q-modification levels in individual tRNAs from human cells based on the presence of a cis-diol in the Q modification. This cis-diol moiety slows modified tRNA migration through polyacrylamide gels supplemented with N-acryloyl-3-aminophenylboronic acid (APB) compared to the unmodified tRNA. This difference can be visualized by Northern blots using probes for specific tRNA.
Keywords: QueuosineBackground
Transfer RNAs (tRNA) contain nucleotide modifications that perform a wide range of roles in translation and in the generation of tRNA fragments that are functional small RNAs. Queuosine (Q, Figure 1) is a tRNA modification that occurs in the wobble anticodon position of tRNAs coding for amino acids Tyr, His, Asn, and Asp. All these tRNAs contain G34U35N36 anticodon sequences. In bacteria, G34-to-Q34 modification can be synthesized de novo through a cascade of enzyme reactions (El Yacoubi et al., 2012). In mammals, the G34 base in these tRNAs can be replaced with queuine (Figure 1) derived from either gut microbiome or diet by a two-protein complex enzyme QTRT1 (catalytic subunit) and QTRT2 encoded in the host genome. Q is widely distributed among prokaryotic and eukaryotic organisms, but its cellular roles are still not fully understood (Rakovich et al., 2011; Fergus et al., 2015; Tuorto et al., 2018; Wang et al., 2018). For example, tRNA Q-modification levels have been correlated with cancer progression and cellular metabolism.
To understand the role of Q-tRNA modification in mammalian cells, we performed comparative studies of cells having various levels of Q-modification (Wang et al., 2018). We found that tRNAs in cultured cell lines are Q-modified at 5%-60% in standard culture medium. Using dialyzed serum medium which was deprived of small molecules like queuine and continued passage of cell divisions, tRNA Q-modification levels continuously decreased until they became undetectable (0Q cells). The addition of 0.1-1 µM queuine to the media restored Q-modification to 100% levels within 12-24 h (100Q cells). To measure the level of tRNA Q-modification in individual tRNA, we used a previously developed method on the basis of the structure of queuosine (Igloi and Kossel, 1985; Zaborske et al., 2014). Cis-diol moieties, such as the 3’ ribose of a tRNA, slow migration through polyacrylamide gels supplemented with N-acryloyl-3-aminophenylboronic acid (APB). Since queuosine has an additional ribose moiety, the migration of Q-modified tRNA in APB gel is slower compared to unmodified tRNA. This differential migration of Q-modified versus unmodified tRNA can be determined by Northern blot analysis using probes specific for each tRNA. The mammalian tRNAAsp and tRNATyr are further glycosylated at the cis-diol of the modified base with mannose (man-Q) or galactose (gal-Q) which prevents the reaction between the Q-base and APB; our approach therefore only works for tRNAHis and tRNAAsn in mammalian cells. Using this method and other techniques, we showed that Q-modification protects cognate tRNAs against ribonuclease cleavage in vitro and in cells which alters the abundance of tRNA fragments (tRF) in human cells (Wang et al., 2018).
Previously, the Q-content in total tRNA in mammalian cells was evaluated by taking advantage of the irreversible nature of the N-glycosidic bond of the Q-base to the sugar-ribose backbone. Total tRNAs were isolated and the extent of Q-modified in total tRNA was measured by the exchange of 3H-guanine with the guanine base of unmodified tRNAs catalyzed by the E. coli TGT enzyme. This exchange reaction was blocked by the Q-modification of the same tRNAs. Total Q-content was estimated based on the fraction of tRNAs that did not show exchange. This approach was first used to determine the Q-modification content in mammalian tissues and bacterial cells during different stages of development and aging (Singhal et al., 1981). Although useful, this previous method could not measure Q-modification level of individual tRNA. In contrast, our APB-gel based method directly measures the Q-modification levels of individual tRNA species starting with as little as 1-2 µg of total RNA.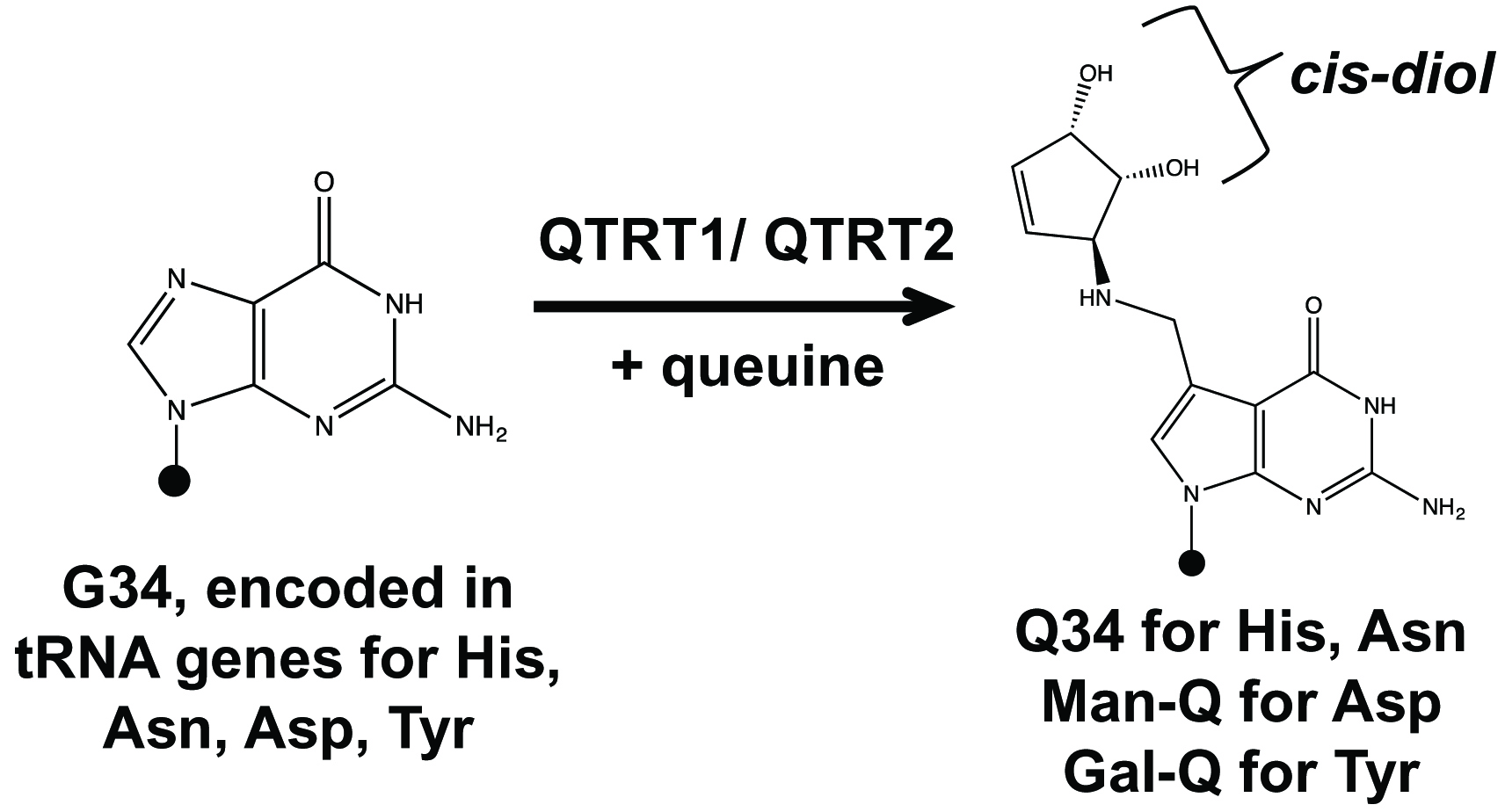
Figure 1. Queuosine (Q) tRNA modification in four tRNAs. Diagram of G34 to Q34 modifications in the wobble anticodon position of tRNAHis/Asn/Asp/Tyr. Queuine corresponds to the nucleobase of Q-nucleotide. QTRT1 and QTRT2 (formerly QTRTD1) proteins are encoded in the human genome. Black dots represent the tRNA body; the QTRT1/QTRT2 complex introduces Q modification in the place of G using queuine as substrate. Q34 in tRNAAsp and tRNATyr are further glycosylated at the cis-diol of the modified base with mannose (man-Q) or galactose (gal-Q) in mammals. Our method measures tRNAHis and tRNAAsn, but not tRNAAsp and tRNATyr because of their glycosylation.
Materials and Reagents
Note: All reagents were purchased from Thermo Fisher if not indicated otherwise and were stored at room temperature.
- Pipette tips
- Plastic foil
- Syringe
- Gel spacers 0.8 mm (Cole-Parmer, UX-28573-12)
- Hybridization tubes 2 x 200 mm (VWR, catalog number: 32645-030)
- Whatman paper (WhatmanTM, 10427812 Grade)
- Hybond-XL membrane (GE Healthcare, catalog number: 45-001-148)
- 5’ 32P-labeled oligonucleotide probes (see Procedure B)
- [γ-32P] ATP (PerkinElmer, catalog number: NEG035C005MC) (storage: -20 °C)
- T4 Polynucleotide kinase (10 U/µl) (Thermo Fisher, catalog number: EK0032) (storage: -20 °C)
- 1x T4 PNK buffer
- 3-Acrylamidophenylboronic acid (Frontier Scientific, catalog number: A10094-5g) (storage: 4 °C)
- 40% acrylamide/bis solution, 29:1 (Bio-Rad, catalog number: 161-0146)
- Acetic acid
- DMEM (Thermo Fisher, catalog number: 11965092)
- EDTA
- Ethanol, 95.5% USP (Ricca, catalog number: R2916000-1A)
- Fetal Bovine Serum dialyzed (Thermo Fisher, catalog number: 26400044)
- Queuine Hydrochloride (Toronto Research Chemicals, catalog number: Q525000)
- Sodium acetate
- Sodium chloride (NaCl)
- Sodium dodecyl sulfate (SDS)
- Sodium phosphate monobasic (NaH2PO4) (Thermo Fisher, catalog number: BP329)
- Sodium phosphate dibasic (Na2HPO4) (Thermo Fisher, catalog number: S374)
- 10x TAE buffer (Sigma-Aldrich, catalog number: 11666690001)
- Tris (Bioland Scientific, catalog number: CT01-5kg)
- Urea
- Bromophenol blue
- Xylene cyanol
- 10x TBE buffer (EMD Millipore, catalog number: 8850-OP)
- Sodium acetate (KOAc)
- Sodium chloride (KCl)
- Nuclease-free water
- 2x Gel loading buffer (see Recipes)
- 10% denaturing polyacrylamide gel (see Recipes)
- 10% APB polyacrylamide gel (see Recipes)
- Dialyzed serum medium (see Recipes)
- Gel extraction buffer (see Recipes)
- Hybridization buffer (see Recipes)
- 1 M Sodium phosphate buffer (pH 7.0) (see Recipes)
- 1 M Tris-HCl buffer (pH 9.0) (see Recipes)
- Washing solution (see Recipes)
Equipment
- Pipettes
- Microcentrifuge (Eppendorf, model: 5415D)
- Hybridization Oven (UVP, model: 95-0030-01)
- Phosphoimager (Bio-Rad, Molecular Imager Pharosfx Plus System, 170-9460)
- Phosphor screens (Bio-Rad or Fuji)
- X-ray Cassette K 35 x 43 cm (Bio-Rad)
- Polyacrylamide gel electrophoresis apparatus with glass plates 17 x 15 cm (Biometra, Model V16-2)
- Thermoblock 24 x 1.5 ml (Eppendorf, 022670522)
- Tube rotator (Cole-Parmer Ltd, model: SB3)
- Vacuum gel dryer (Bio-Rad, Model 583 Gel Dryers)
- Incubator
- 4 °C refrigerator
- -20 °C freezer
Software
- Quantity One (Bio-Rad Laboratories, Inc., www.bio-rad.com/)
Procedure
文章信息
版权信息
© 2019 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Matuszek, Z. and Pan, T. (2019). Quantification of Queuosine Modification Levels in tRNA from Human Cells Using APB Gel and Northern Blot. Bio-protocol 9(6): e3191. DOI: 10.21769/BioProtoc.3191.
分类
微生物学 > 微生物生物化学 > RNA
分子生物学 > RNA > RNA 检测
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link