- EN - English
- CN - 中文
QsRNA-seq: A protocol for Generating Libraries for High-throughput Sequencing of Small RNAs
QsRNA-seq: 小RNA高通量测序文库构建方案
发布: 2019年03月05日第9卷第5期 DOI: 10.21769/BioProtoc.3179 浏览次数: 8226
评审: Gal HaimovichOmar AkilAnonymous reviewer(s)
Abstract
Small RNAs (sRNAs) are 20-30 nt long non-coding RNA molecules that regulate essentially all cellular processes. Besides being an intensively studied topic in academic research, sRNAs also hold a promise as clinical biomarkers. While the need for expressional profiling of sRNAs is growing, preparation of sRNA libraries for high-throughput sequencing (HTS) remains technically challenging, due to their small size. The common PAGE-based protocol is time-consuming and inefficient due to material loss, while gel-free protocols generate libraries of insufficient quality. To overcome these shortcomings, we modified the conditions of size-selection by Solid Phase Reversible Immobilization (SPRI) in a way that allows separation of nucleic acids shorter than 100 nt and differing in length by only 20 nt. Implementing the method for preparation of small RNA libraries for HTS resulted in QsRNA-seq, a gel-free, fast and easy-to-perform protocol, amenable to automation, generating very clean libraries that result in high-depth expression data. The protocol also utilizes Unique Molecular Identifiers (UMI) for reduction of library preparation biases and to quantify expression levels. QsRNA-seq provides an excellent solution to the growing needs for small RNA expression profiling for research clinical use.
Keywords: Small RNA (小RNA)Background
Although small RNAs, which are 20-30 nt long regulatory non-coding RNA molecules, constitute a tiny percentage of the total cellular RNA, they have a huge impact on many cellular processes (including, gene regulation, organism development, adaptation to changing environment and so on) (Jost et al., 2011). There are several classes of small RNA (Youngman and Claycomb, 2014) with microRNAs (miRNAs) being the most extensively studied (Bartel, 2009).
Besides being of a great interest for basic research, miRNAs hold promise as diagnostic markers. Due to their exceptional stability, intact miRNAs are present in preserved tumor samples (formalin-fixed paraffin-embedded [FFPE]), which exhibit a near total mRNA degradation (Nagy et al., 2016). Moreover, stable miRNAs circulate in body fluids such as plasma, urine and sputum of healthy and diseased patients (Mitchell et al., 2008). Liquid biopsy of circulating miRNAs can serve as potential markers for a broad range of human diseases especially for diseases for which ordinary biopsies cannot be obtained, such as cardiovascular diseases, and psychiatric diseases (Kim, 2015; Backes et al., 2016).
While assessing the repertoire and abundance of cellular miRNAs became an important aspect in many studies, the tools to do so lag behind. High-throughput sequencing (HTS) is a gold standard method for mRNA profiling, extensively used in both basic research and clinical settings. In striking contrast, HTS profiling of miRNAs is not widespread, as preparation of small RNA libraries remains technically challenging.
To become compatible with Illumina or other HTS platforms, small RNAs are ligated from both sides (5’ and 3’) to oligonucleotide adapters and then reverse transcribed. The resulting small RNA library is then amplified by PCR with primers complementary to the 5’ and 3’ adapters that include sequences recognized by the sequencing machinery (Lu et al., 2007). If small RNAs are not separated from other RNA species, in particular from tRNA, which are highly abundant and close in size (~70 bp), the resulting libraries will be highly contaminated by irrelevant sequences. In addition, 3’ and 5’-adapters ligate to each other, forming adapter-dimers, which contaminate libraries with null sequences. Thus, for generation of a high-quality sRNA library, a preparation protocol must enable separation of small RNA from total RNA and separation of the ligation products from free adapters. Size-selection on Solid Phase Reversible Immobilization (SPRI) paramagnetic beads (Lundin et al., 2010), used in preparation of mRNA and DNA libraries, was not suitable for preparation of small RNA libraries, as the lower limit of size discrimination is 100 nt (Paithankar et al., 1991). The only remaining option, electrophoresis on PAGE gel, is cumbersome, time-consuming, requires technical skills, results in a huge loss of the product and eliminates the possibility of automatization. Preventing adapter-dimer contamination by molecular biology strategies (Kawano et al., 2010; Vigneault et al., 2012) is not a good alternative to size-selection as it does not solve the problem of contamination by irrelevant sequences.
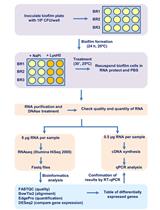
To allow easy size selection of short nucleic acid molecules, we modified the SPRI method by using a combination of two crowding agents, Polyethylene Glycol (PEG) and Isopropanol. This modification enables separation of nucleic acid fragments shorter than 100 nt, differing in length by only 20 nt (Fishman et al., 2018). Exploiting this new separation method, we designed a protocol for preparation of small RNA libraries for high-throughput sequencing based on Lau et al. (2001) and we named the protocol QsRNA-seq. The input material for the protocol is either total RNA or low molecular weight RNA fraction (LMW RNA, < 200 nt). During the protocol three SPRI-based size selection steps are performed, each using different conditions, to allow separation of sRNA from total RNA and of ligation products from free adapters after each ligation (for the protocol outline see the Procedure section below). In addition, the protocol supports the possibility of multiplexing samples by adding a 4-nt barcode to the 5’-adapter. The protocol also supports using unique molecular identifiers (UMI) for reduction of ligation bias (Fuchs et al., 2015) and correction of amplification bias (Kivioja et al., 2011) by adding an 8-nt random sequence to the 5’-adapter as well. To correct the amplification bias, identical small RNA sequences with the same UMI are considered to be amplification products and merged to one sequence (i.e., collapsed) during data processing. By comparing libraries generated from the same RNA using adapters with the same barcode either with UMI or without UMI, we found that adding UMI significantly reduced both biases (Fishman et al., 2018).
QsRNA-seq has several significant advantages over the existing protocols. The entire protocol can be performed in one day and does not require high manual skills. Because it does not require any gel use there is less material lost, which enables small quantities of the input RNA to be analyzed. Moreover, it allows simultaneous processing of many samples as well as automation, turning the preparation of small RNA libraries into a routine procedure. In addition, usage of UMI allows reduction of library preparation biases, resulting in more reliable quantitation. To summarize, QsRNA-seq is an easy-to-use protocol that generates reliable and quantifiable high-throughput small RNA libraries.
Materials and Reagents
- RNase-free plastic pipette tips with filter (10 μl, 20 μl, 200 μl, 1,000 μl)
- 1.7 ml RNase-free microtubes
- 0.2 ml RNase-free PCR tubes
- miRVana miRNA Isolation Kit (Thermo Fisher Scientific, catalog number: AM1560) or TRIzol reagent (Ambion, catalog number: 15596026) with Direct-Zol RNA MiniPrep Plus (Zymo Research, catalog number: R2072)
- RiboLock RNase Inhibitor (Thermo Fisher Scientific, catalog number: EO0381)
- T4 RNA ligase 1 (ssRNA Ligase) (NEB, catalog number: M0204S)
- T4 RNA ligase 2, truncated (NEB, catalog number: M0242S)
- RppH enzyme (NEB, catalog number M0356S)
- QScript Flex cDNA synthesis kit (Quanta, catalog number: 95049-025)
- Phusion High-Fidelity DNA Polymerase (NEB, catalog number: M0530S)
- Agencourt Ampure XP (Beckman Coulter, catalog number: A63881) or SPRI select reagent Kit (Beckman Coulter, catalog number: B23319)
- RNA BR assay kit (Quibit, catalog number: Q10210)
- dsDNA HS assay kit (Quibit, catalog number: Q10210)
- PEG 8000 (NEB, catalog number: B1004)
- Nuclease-free water (Sigma-Aldrich, catalog number: W4502)
- Ethyl Alcohol absolute
- Isopropyl-alcohol (Isopropanol) chemical grade
- Ethidium Bromide (hylabs, catalog number: BP451)
- Gel loading dye purple (6x) (NEB, catalog number: B7024)
- [Optional] Low-range ultra agarose (Bio-Rad, catalog number: 161-3107)
- ATP (NEB, catalog number: P0756S)
- DMSO (NEB, catalog number B0515)
- Tris (Spectrum, catalog number: s1519)
- Boric Acid (Bio-Lab, catalog number: 000201059100)
- EDTA (J.T. Baker, catalog number: 8993-01)
- 50 bp DNA ladder (NEB, catalog number: N3236s)
- Oligonucleotides: all the oligonucleotides were ordered from IDT (www.idtdna.com)
- [Optional] NucleoSpin gel and PCR Clean-up kit (Macherey-Nagel, catalog number: 740609.50) or Gel/PCR DNA fragments Extraction kit (Geneaid, catalog number: DF004)
- 5x TBE (see Recipes)
Equipment
- Micro-pipettors (Gilson, Pipetman)
- DynaMag-2 magnetic stand (Thermo Fisher Scientific, catalog number: 12321D)
- Benchtop microcentrifuge, non-refrigerated (5424 with Rotor FA-45-24-11, Eppendorf, catalog number: 5424000010)
- Vortex mixer, flat head, Vortex-Genie 2 (Scientific Industries, model: SI-0266)
- Cooling block for 1.7 ml tubes (MS, catalog number: MC-0203)
- Agarose gel electrophoresis apparatus
- Electrophoresis power supply
- Gel imager (Bio-Rad, Gel Doc XR+)
- Fluorometer, Quibit 2.0 (Invitrogen, model: Q32866)
- Thermocycler (Biometra, TProfessional)
- Tapestation/Bioanalyzer
Procedure
文章信息
版权信息
© 2019 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Fishman, A. and Lamm, A. T. (2019). QsRNA-seq: A protocol for Generating Libraries for High-throughput Sequencing of Small RNAs. Bio-protocol 9(5): e3179. DOI: 10.21769/BioProtoc.3179.
分类
分子生物学 > RNA > RNA 测序
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link