- EN - English
- CN - 中文
flySAM Transgenic CRISPRa System Manual
基于flySAM的CRISPRa转基因系统操作手册
发布: 2019年01月20日第9卷第2期 DOI: 10.21769/BioProtoc.3147 浏览次数: 9614
评审: Gal HaimovichAbhijit KaleAnonymous reviewer(s)
Abstract
Powerful and general methods that can enhance gene expression are useful to systematically study gene function. To date, compared with the methods in generating loss-of-function mutants, methods to achieve gain-of-function are limited. The entire field in Drosophila has relied heavily on the Gal4/UAS:cDNA overexpression system developed over two decades ago. It is laborious and expensive to clone the coding DNA sequence (CDS) of a gene, especially those of large size. In addition, side effects of this method are often observed because of the ectopic expression. Also, simultaneous activation of two genes with the traditional method is often time-consuming, and few are achievable for three or more genes. In this protocol, we describe how to build an effective and convenient targeting activator system, flySAM, to activate endogenous genes in Drosophila melanogaster based on the structure-guided engineering of CRISPR-Cas9 complex.
Keywords: CRISPR-Cas nuclease (CRISPR-Cas核酸酶)Background
Currently, most of the genes in Drosophila can be knocked down or knocked out through powerful genetic tools and several loss-of-function (LOF) resources have been established based on these tools (Ren et al., 2013; Qiao et al., 2018). A similar gain-of-function (GOF) resource is valuable to study gene function as it can be a complement to LOF. However, we do not have a genome-wide GOF resource until now mainly because of technical difficulties. The traditional gene overexpression system in Drosophila is the Gal4/UAS:cDNA system. It needs to clone the coding DNA sequence (CDS) of a gene downstream of the upstream activation sequence (UAS) which can be controlled by Gal4 transcription factor. It is laborious, expensive and sometimes unachievable. Therefore, an easier way to realize GOF is urgently needed. The clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) system is an effective genome-editing tool. The transcriptional activation system in Drosophila based on the CRISPR/Cas9 technique, termed “CRISPR transcriptional activation (CRISPRa) system”, has been developed in recent years (Konermann et al., 2015; Lin et al., 2015; Ewen-Campen et al., 2017). Cas9 has HNH and RuvC nuclease domains which are responsible for the cutting of complementary and non-complementary DNA strand, respectively. Through introducing single amino acid mutation within HNH and RuvC domains of Cas9 (dCas9), nuclease activity of Cas9 is abolished without affecting its DNA binding ability (Dominguez et al., 2016). The CRISPR transcriptional activation CRISPRa system is generated by fusing several activation domains to dCas9 and can be recruited to the upstream of the Transcription Start Site (TSS) through single-guide RNAs (sgRNAs). The specificity of CRISPRa system is controlled by 20 bp guide sequence in sgRNA, thus one can simply use the 20 bp guide sequence against different targets. Previously, VPR-based CRISPRa system has been built in Drosophila in vivo and in vitro (Lin et al., 2015; Ewen-Campen et al., 2017). In this system, three activation domains, VP64, P65, Rta (VPR), tandemly following the dCas9. However, due to low efficiency, this system relies on two sgRNAs to boost gene transcription, which not only increased the complicacy and cost of the cloning procedure, but also increased toxicity and the chances of off-target effects. In addition, although it works both in vitro and in vivo, it often produces weaker phenotypes than the traditional Gal4/UAS:cDNA system.
To improve the efficiency, reduce the toxicity, and streamline the cloning procedure, this protocol introduces how to construct transcriptional activation lines based on flySAM system. In this system, the dCas9-VP64 and MCP-p65-HSF1 is controlled by 10x UAS and sgRNA2.0 which is driven by U6:2 promoter (Jia et al., 2018). Unlike the previous methods, single sgRNA can successfully activate gene transcription. In addition, to study the intrinsic and extrinsic roles of genes with redundant functions or involved in protein complex, we can easily clone multiple sgRNAs to target different genes.
Materials and Reagents
- 1.5 ml MaxyClear Microtubes (Axygen, catalog number: MCT-150-C)
- 0.2 ml Polypropylene PCR Tube (Axygen, catalog number: PCR-0208-C)
- Pipette tips (Corning, Axygen®)
- 0.22 μm Millipore filters (Millipore, catalog number: R7MA69670)
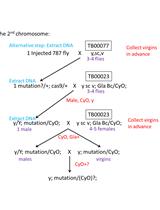
- Fly stocks (All Drosophila strains are stored in Tsinghua Fly Center)
- y sc v nanos-integrase; attP40 (TB00016)
- y sc v nanos-integrase;; attP2 (TB00018)
- y sc v (TB00077)
- y sc v; Gla Bc/CyO (TB00023)
- y sc v;; Dr,e/TM3,Sb (TB00139)
- Trans5α Chemically Competent Cell (Transgen Biotech, catalog number: CD201-01)
- flySAM (Ampicillin resistant, plasmid map is shown in Figure 1)
- BbsI (New England Biolabs, catalog number: R0539L)
- NheI (New England Biolabs, catalog number: R3131L)
- SpeI (New England Biolabs, catalog number: R3133L)
- T4 DNA ligase (New England Biolabs, catalog number: M0202L)
- Alkaline phosphatase, calf intestinal (CIP) (New England Biolabs, catalog number: M0290S)
- AxyPrep Plasmid Miniprep Kit (AXYGEN, catalog number: AP-MN-P-250)
- AxyPrep DNA Gel Extraction Kit (AXYGEN, catalog number: AP-GX-250)
- PurePlasmid Mini Kit (CWBiotech, catalog number: CW0500M)
- Forward Primer 1: 5’-TCAACAAACGaacaataggacac-3’
- Reverse Primer 1: 5’-aAAAAAGCACCGACTCGGTG-3’
- Forward Primer 2: 5’-ACATCAGGAAAGAGCAGTTGAG-3’
- Reverse Primer 2: 5’-TTGCTCACCTGTGATTGCTCC-3’
- 50x TAE (Double Helix, catalog number: P0309A)
- Agarose (Invitrogen, catalog number: 0000602315)
- LB powder (BD, catalog number: 2171199)
- KCl (AMRESCO, catalog number: 7447-40-7)
- Tris-HCl (AMRESCO, catalog number: 1185-53-1)
- EDTA (AMRESCO, catalog number: 60-00-4)
- NaCl (AMRESCO, catalog number: 7647-14-5)
- Ampicillin (Solarbio, catalog number: A8180)
- Ethidium bromide (Sigma, catalog number: E8751)
- GoTag Green Master Mix, 2x (Promega, catalog number: 000179370)
- Sodium phosphate buffer (see Recipes)
- 10x Injection buffer (see Recipes)
- 10x Annealing buffer (see Recipes)
- LB (Luria Bertani) medium (see Recipes)
- 1x TAE (see Recipes)
- 1.5% agarose gel (see Recipes)
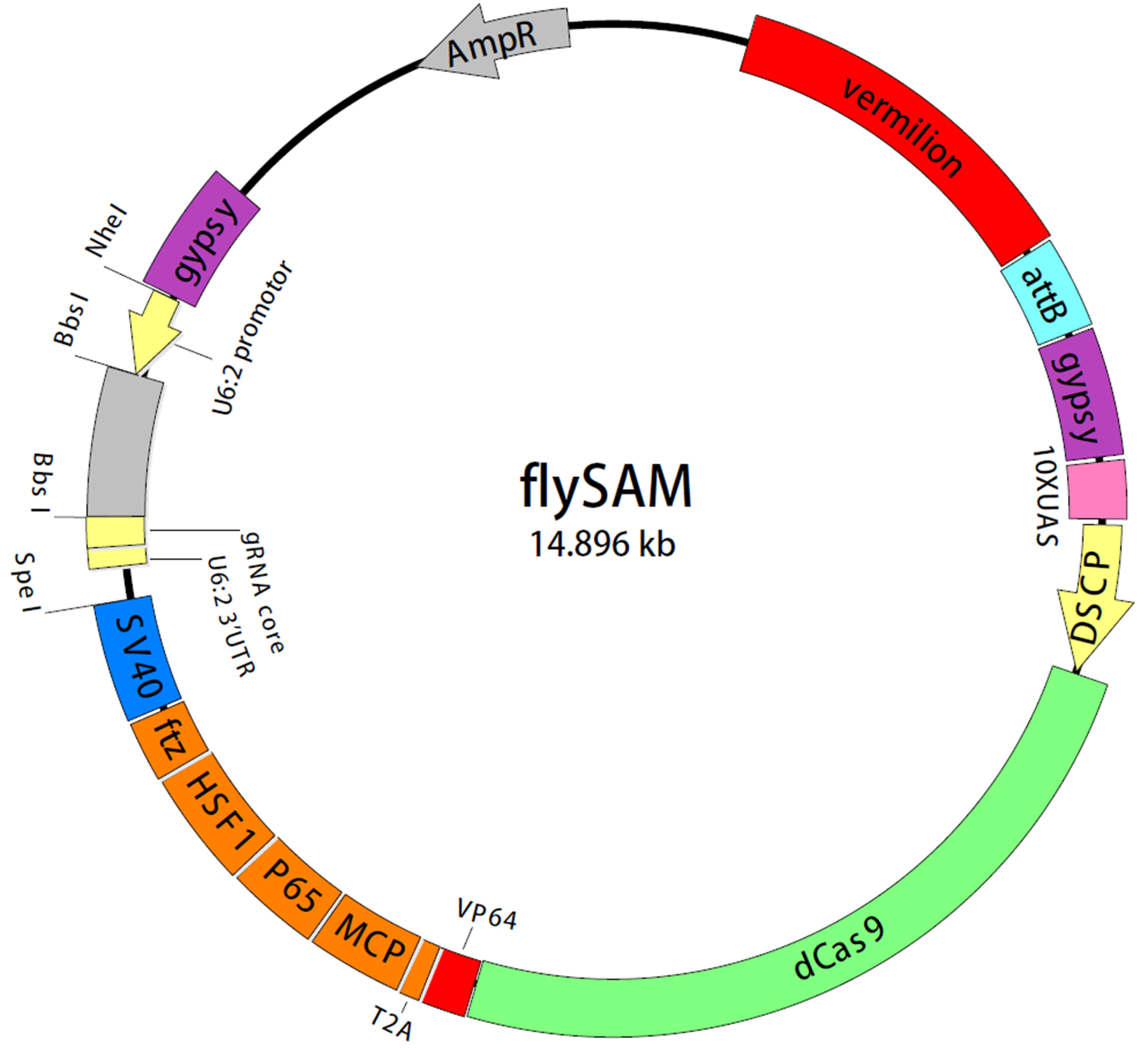
Figure 1. The map of flySAM construct. The dCas9 is under the control of 10 x UAS and Drosophila synthetic core promoter (DSCP) and followed by VP64. dCas9-VP64 and MCP-P65-HSF1 are mediated and separated by T2A self-cleaving peptide. ftz intron is located between CRISPRa components and SV40 polyA tail. The grey block between two BbsI sites is a marker to detect whether the sgRNA is under the control of U6:2 regulatory sequence. Two gypsy elements which can boost gene expression are used to flank the CRISPRa components. The attB sequence and a vermillion+ marker are also included in this plasmid.
Equipment
- Pipette (Eppendorf)
- Microwave (SANYO, model: EM-2509EB1)
- Autoclave (SANYO, model: MLS-3780)
- PCR Thermal Cycler (Eppendorf, Mastercycler nexus GSX1)
- Microcentrifuge (Eppendorf, model: 5417R)
- DNA electrophoresis apparatus (Bio-Rad)
- NanoDrop 2000 Spectrophotometer (Thermo Scientific)
Procedure
文章信息
版权信息
© 2019 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Jia, Y., Shen, D., Wang, X., Sun, J., Peng, P., Xu, R., Xu, B. and Ni, J. (2019). flySAM Transgenic CRISPRa System Manual. Bio-protocol 9(2): e3147. DOI: 10.21769/BioProtoc.3147.
分类
分子生物学 > DNA > 基因表达
分子生物学 > DNA > DNA 修饰
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link