- EN - English
- CN - 中文
Variable Dose Analysis: A Novel High-throughput RNAi Screening Method for Drosophila Cells
可变剂量分析:一种果蝇细胞高通量RNAi筛选的新方法
发布: 2018年12月20日第8卷第24期 DOI: 10.21769/BioProtoc.3112 浏览次数: 5354
评审: Tzvetina BrumbarovaPradeep Kumar BhaskarAnonymous reviewer(s)
Abstract
Genetic screens are a powerful approach to identify previously uncharacterized genes involved in specific biological processes. Several technologies have been developed for high-throughput screens using reagents such as RNAi or CRISPR, and each approach is associated with specific advantages and disadvantages. Variable Dose Analysis (VDA), is an RNAi-based method developed in Drosophila cells that improves signal-to-noise ratio compared to previous methods. VDA assays are performed by co-transfecting cells with a plasmid expressing shRNA, (a type of RNAi that can be easily expressed from a DNA plasmid) against a gene of interest and a second plasmid expressing a fluorescent reporter protein. Fluorescent protein expression, can be used as an indirect readout of shRNA expression and therefore target gene knockdown efficiency. Using this approach, we can measure phenotypes over a range of knockdown efficiencies in a single sample. When applied to genetic interaction screens, VDA results in improved consistency between screens and reliable detection of known interactions. Furthermore, because phenotypes are analyzed over a range of target gene knockdown efficiencies, VDA allows the detection of phenotypes and genetic interactions involving essential genes at sub-lethal knockdown efficiency. This therefore represents a powerful approach to high-throughput screening applicable to a wide range of biological questions.
Keywords: RNAi (RNAi)Background
High throughput screens (HTS) are experiments in which the functions of many individual genes are systematically disrupted one at a time and the resulting phenotypes measured. Screens are generally performed by treating cells in culture with genetic or pharmacological reagents targeting individual gene functions and measuring the effects of these treatments on a phenotype of interest (Mohr et al., 2014). Using this approach, it is possible to identify genes and pathways that function in a specific biological process. Alternatively, screens can be performed in healthy and disease model cell lines and results compared to study the mechanism of the disease or identify candidate therapeutic targets (Zhan and Boutros, 2016).
In addition to ‘single gene’ screens, it is possible to perform genetic interaction screens, in which pairs of genes are targeted, and assess how phenotypes vary compared to what is expected based on disruption of either gene alone. This approach can be used to map genetic interactions and gain insights into the structure and function of biological pathways and networks (Tong et al., 2004; Fischer et al., 2015).
In Drosophila cells, genetic screens are often performed using long double stranded RNA (dsRNA) reagents. Simply by bathing cells in culture media containing dsRNA, the reagent is efficiently taken up and processed to inhibit the function of the target gene. Given the simplicity of this system, it is possible to perform screens on a genome scale with relative ease. However, there are a number of limitations associated with this approach. For example, background noise as well as incomplete or variable disruption of the target gene may result in a failure to detect all but the strongest phenotypes (Boutros and Ahringer, 2008). Moreover, false positives can occur when off-target effects lead to disruption of unintended genes (Ma et al., 2006). Such limitations are even more problematic in genetic interaction screens when two genes must be efficiently and specifically inhibited at once.
Variable dose analysis (VDA) is a novel approach to RNAi assays in which each cell within a population receives a different dose of short hairpin RNA (shRNA) (Housden et al., 2017). Using a fluorescent reporter, the relative knockdown efficiency in each cell can be measured, and the relationship between the efficiency of target-gene disruption and a feature of interest analyzed in order to detect phenotypes (Figure 1A). This differs from previous methods, which typically measure the average phenotype over a population of cells and results in approximately a 2.5-fold increase in signal-to-noise ratio compared to standard dsRNA methods. In addition, VDA facilitates the study of essential genes because phenotypes can be measured at sub-lethal knockdown efficiency.
While VDA represents an improved approach to HTS in many ways, there are a number of disadvantages to this method and continued development will be necessary to remove these limitations (Figure 1B). For example, VDA requires the use of transfection, which increases reagent costs and reduces throughput compared to dsRNA-based methods. In addition, there are currently relatively few reagent libraries compatible with VDA although additional libraries are under construction. Finally, VDA may not be applicable to all cell types due to the need for efficient and variable transfection efficiency. Thus, cell types that are not easily transfected cannot be studied using this method. Nevertheless, although VDA has so far only been applied in Drosophila cells, it can in principle be applied to a wide variety of phenotypic readouts and cell types, making it an attractive alternative tool for HTS.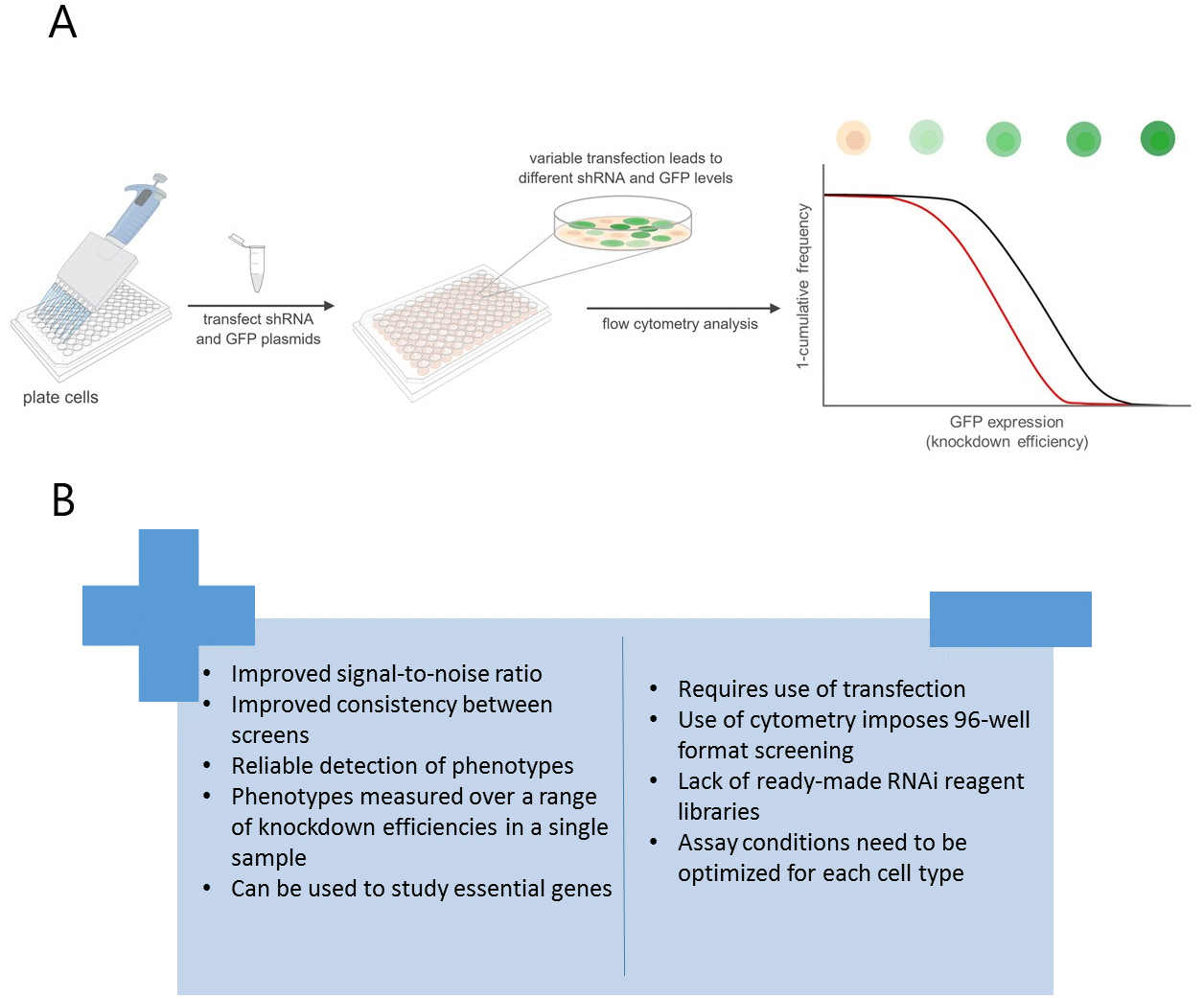
Figure 1. VDA provides improvement over dsRNA-based methods. A. Schematic depicting the VDA HTS pipeline. Control shRNA in black, lethal shRNA in red. B. Advantages and disadvantages of VDA compared to dsRNA screens.
Materials and Reagents
- Pipette tips (Gilson, catalog numbers: F167101, F167103) (storage conditions: RT)
- NuncTM Cell Culture Treated flasks with filter caps (Thermo Scientific, catalog number: 136196) (storage conditions: RT)
- 96-well tissue culture plate (JET Biofil, catalog number: TCP011096) (storage conditions: RT)
- 96-well PCR plate (BRAND, catalog number: 781375) (storage conditions: RT)
- Parafilm M (Bemis, catalog number: HS234526A) (storage conditions: RT)
- Paper towel (any brand)
- Drosophila S2R+ cells (DGRC, catalog number: 150) (storage conditions: 25 °C)
- FuGENE® HD transfection reagent (Promega, catalog number: E2311) (storage conditions: 4 °C)
- Schneider’s Drosophila Medium (Gibco, catalog number: 21720024) (storage conditions: 4 °C)
- One ShotTM Fetal Bovine Serum (Gibco, catalog number: A3382001) (storage conditions: -20 °C)
- Penicillin-Streptomycin (Gibco, catalog number: 15070063) (storage conditions: -20 °C)
- ^pActin-GFP (storage conditions: -20 °C)
- ^pActin-GAl4 (storage conditions: -20 °C)
- *shRNA library (storage conditions: -20 °C)
- Phosphate Buffered Saline (PBS) (Gibco, catalog number: 20012019) (storage conditions: RT)
- Culture media (see Recipes)
Notes:
- ^Available from the Housden lab.
- *Available from the DRSC (fgr.hms.harvard.edu) or Housden lab.
- RT: Room temperature.
Equipment
- Flow cytometer (Beckman Coulter, model: CytoFlex S)
- 25 °C incubator (any supplier)
- Tissue culture hood (any supplier)
- Multichannel pipettes (Gilson, model: PIPETMAN®)
- Humidifying chamber (N/A)
Software
- Matlab (MathWorks)
- Excel (Microsoft)
Procedure
文章信息
版权信息
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Sierzputowska, K., Baxter, C. R. and Housden, B. E. (2018). Variable Dose Analysis: A Novel High-throughput RNAi Screening Method for Drosophila Cells. Bio-protocol 8(24): e3112. DOI: 10.21769/BioProtoc.3112.
分类
分子生物学 > RNA > RNA 干扰
细胞生物学 > 基于细胞的分析方法 > 流式细胞术
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link