- EN - English
- CN - 中文
A Functionally Robust Phenotypic Screen that Identifies Drug Resistance-associated Genes Using 3D Cell Culture
利用3D细胞培养鉴定耐药相关基因的一种功能性稳健表型筛选
发布: 2018年11月20日第8卷第22期 DOI: 10.21769/BioProtoc.3083 浏览次数: 7353
评审: Nicoletta CordaniSophie A LelièvreAnonymous reviewer(s)
Abstract
Drug resistance is a major obstacle in cancer treatment: A case in point is the failure of cancer patients to respond to tyrosine kinase inhibitors (TKI) of EGFR, a receptor that is highly expressed in many cancers. Identification of the targets and delineation of mechanisms of drug resistance remain major challenges. Traditional pharmacological assays of drug resistance measure the response of tumor cells using cell proliferation or cell death as readouts. These assays performed using traditional plastic tissue culture plates (2D) do not translate to in vivo realities. Here, we describe a genetic screen based on phenotypic changes that can be completed over a period of 1-1½ months using functional endpoints in physiologically relevant 3D culture models. This phenotype-based assay could lead to the discovery of previously unknown therapeutic targets and could explain the source of the resistance and relapse. As a proof of principle, we performed our 3D culture assay with a small cDNA library in that yielded five unknown intermediates in EGFR and PI3K signaling pathways. Here, we describe the screening method and the characterization of one of the five molecules, but this approach could be easily expanded for a high-throughput screening to identify or evaluate many more unknown intermediates in oncogenic signaling pathways.
Keywords: 3D Culture (3D培养)Background
Despite significant progress in therapeutic strategies, almost 30% of breast cancer patients develop chemotherapy resistance following initial treatment leading to relapse (Brewster et al., 2008; Holohan et al., 2013). Genes or pathways that are responsible for chemoresistance have been identified through traditional screening approaches including gain-of-function and loss-of-function tools (Grimm, 2004; Arnoldo et al., 2014). More recently, a high-throughput screening technique of CRISPR/Cas9-based system has been used to identify molecular targets and their related pathways (Hu and Zhang, 2016; Luo, 2016). Nevertheless, most assays fail to provide a mechanism of resistance and alternative possibilities of treatment.
In a tissue, dynamic and reciprocal signaling between cells and their local microenvironment is required for maintenance of tissue architecture and homeostasis (Bissell and Hines, 2011; Furuta et al., 2018). In particular, biochemical and biophysical cues from the local microenvironment are recognized by cell surface receptors such as growth factor receptors and integrins that initiate a signaling cascade transduced into the nucleus to change gene expression (Bissell et al., 1999; Ghajar and Bissell, 2008). In addition to mutations that drive gene expression involved in drug resistance, the microenvironment plays a significant role in multidrug resistance (Weaver et al., 2002; Correia and Bissell, 2012). Therefore, a physiologically relevant assay system–one that permits screening while recapitulating the microenvironment in vivo–is critical for identifying new targets for breast cancer therapy and for elucidating mechanisms underlying the tissue specificity of drug resistance.
HMT-3522 progression series of human mammary epithelial cells was derived from a reduction mammoplasty of a woman with benign fibrocystic breast disease (Petersen et al., 1990; Briand et al., 1996). The epithelial tissue was cultured in a chemically defined culture medium and the phenotypically normal S1 cells were established (Briand et al., 1987). After over 100 passages of S1 cells in epidermal growth factor-free medium, growth factor-independent and tumorigenic T4-2 cells were obtained (Briand et al., 1996). Non-malignant S1 cells in 3D culture of Laminin-rich extracellular matrix (lrECM) recapitulate an organized polar unit of breast structure referred to as acinus, in culture we refer to them as mammary organoids (Petersen et al., 1992; Weaver et al., 1995; Lee et al., 2007). In the same medium and culture conditions, malignant T4-2 cells form disorganized colonies in 3D culture and tumors in mice. We have shown that malignant T4-2 cells in the presence of lrECM become quiescent and can be reverted to a structure partially resembling non-malignant counterparts, despite still containing the cancer genome (Figure 1). This “phenotypic reversion” is achieved by adjusting extracellular and intracellular signaling pathways, such as inhibitors of receptor tyrosine kinase or the humanized blocking antibody hampering oncogenic signaling which otherwise leads cells to lose architecture and grow out of control (Weaver et al., 1997; Wang et al., 1998; Weaver and Bissell, 1999; Muschler et al., 2002; Wang et al., 2002; Kenny and Bissell, 2007; Itoh et al., 2007; Beliveau et al., 2010; Onodera et al., 2014). Other breast cancer cell lines, such as metastatic MDA-MB-231, can be reverted by treatment with a combination of inhibitors of integrin beta1, PI3K, or MAPK (Wang et al., 2002).
Using the three conditions described above, non-malignant, malignant and reverted malignant cells in our 3D cell culture models, we demonstrated that the efficacy of the chemotherapeutic response was strictly dependent on cell and tissue polarity and tissue architecture (Weaver et al., 2002). By combining the ability to “phenotypically” revert the malignant cells with a gain-of-function genetic screen, we developed a phenotype-based functional genetic screen that allowed the identification of novel genes associated with EGFR -TKI resistance (Lee et al., 2012).
In this protocol, viral cDNA library generated from malignant breast epithelial cells is transduced into the same malignant cells under 2D (tissue culture on plastic) followed by subsequent transfer to treatment with EGFR-TKI (AG1478) or PI3K inhibitor (LY294002), on 3D lrECM culture in the presence of antibiotics for selection of integrated genes. The 3D cultures are maintained for 5-7 days to allow the malignant T4-2 cells to undergo phenotypic reversion in the presence of either inhibitor. Then colonies that fail to revert and continuously grow are isolated from the cultures and subjected to sequencing for identifying the integrated cDNA that conferred resistance to EGFR-TKI or PI3K inhibitor (Figure 2).
Using this technique, we identified five candidates (Table 2) and further characterized one of the genes that conferred EGFR-TKI resistance in breast cancer cells when overexpressed. This gene was identified as FAM83A, one of the eight members of a previously unidentified oncogene family. We observed that FAM83A expression was increased when cells were treated with tyrosine kinase inhibitor, lapatinib; the cells that survived became resistant to the treatment. FAM83A-associated molecular mechanism of drug resistance serves as an example of the utility of this screening method and provides a basis for predictive diagnosis and possible molecular targets for chemotherapy.
This assay can be applied for identifying novel therapeutic targets by modulation of gene expression such as systemic gain-of-function genetic screens with a cDNA library or loss-of-function genetic screens with shRNA (short hairpin RNA) or sgRNA (single guide RNA) libraries. Additionally, this assay is also a useful tool for evaluation of drug response and efficacy of pharmacological candidates in different cancer cell lines allowing us to predict therapeutic outcomes.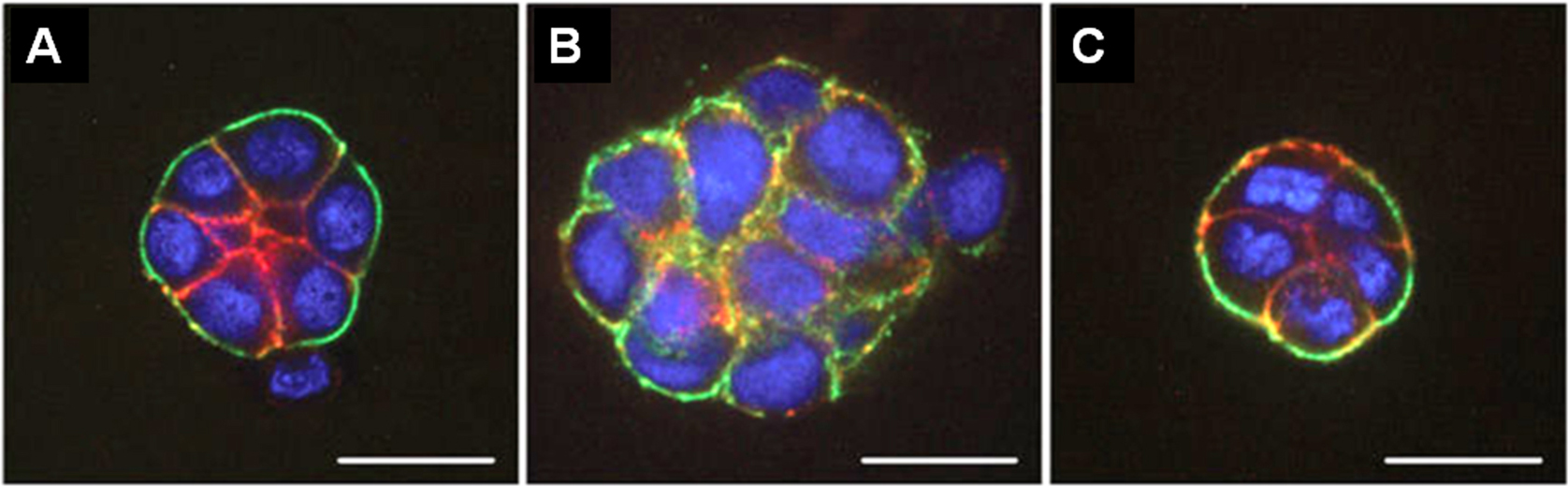
Figure 1. Fluorescence microscopy images of HMT 3522 progression series. A. Non-malignant S1 cells organized into acini structure resembling alveoli in the mammary gland in vivo. B. Malignant T4-2 cells formed large disorganized colonies. C. “Phenotypically reverted” malignant T4-2 cell showed basal polarity and growth arrest by modulation of signal pathways. All cells were grown in 3D culture of lrECM, and stained for α6 integrin (green), β-catenin (red), and nuclei (blue). Adapted from Figure 3 in Lee et al. (2007). 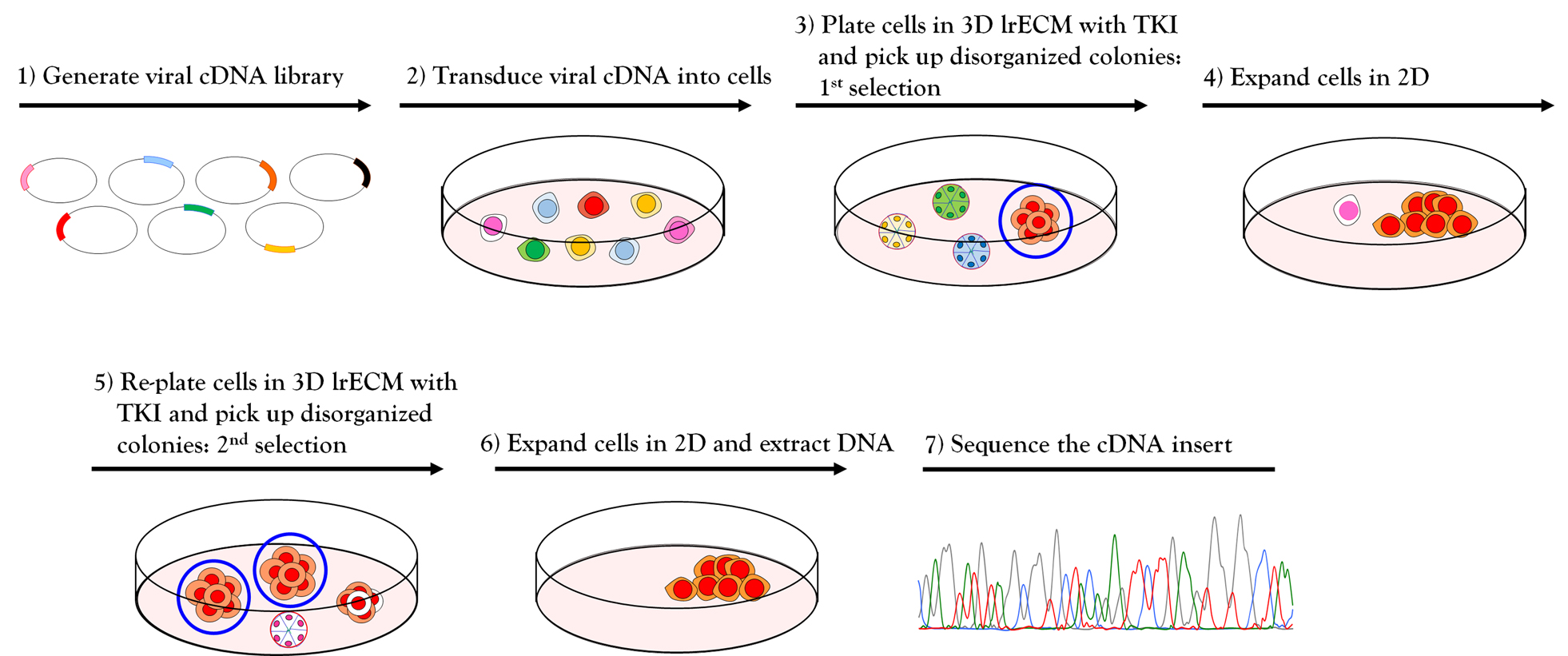
Figure 2. Workflow for phenotype-based genetic screen. (1-2) Amphotropic retroviral cDNA library was generated and transduced into HMT3522 malignant T4-2 cells. (3) The cDNA expressing T4-2 cells were plated on 3D lrECM culture and treated with EGFR-TKI AG1478. Colonies that fail to revert (circled in blue) were isolated from the culture using a flame-drawn glass pipette under a phase-contrast microscope in a Biosafety cabinet. (4) Each isolated colony was transferred to one well of a 48-well tissue culture plate and subsequently expanded to a larger multi-well plate until the cell number reached one million cells. (5) Cells were re-plated on 3D lrECM culture and treated with EGFR-TKI AG1478, and the isolation procedure was repeated as described in (3). (6) Genomic DNA was isolated from selected clones grown in 2D and cDNA inserts were recovered by PCR using the flanking sequences of retroviral vector used for cloning. (7) The amplified PCR products were analyzed for sequencing to determine the identity of inserted cDNA.
Materials and Reagents
- Construction of cDNA library
- Retroviral pESY-Neo vector (used for the construction of cDNA library is a modified version of the pEYK3.1 vector [a kind gift from the laboratory of Dr. George Daley at the Whitehead Institute])
- DNeasy® tissue kit (QIAGEN, catalog number: 69504)
- Oligotex poly (A)+ RNA purification column (QIAGEN, catalog number: 70042)
- SuperScript® plasmid system with Gateway® technology for cDNA synthesis and cloning (Invitrogen Life Technologies, catalog number: 18248013)
- Tris-Borate-EDTA (TBE) buffer (Amresco, 10x liquid concentrate, catalog number: 0658-4l)
- Ultrapure agarose (Invitrogen, catalog number: 16500-100)
- XL-1 Gold Ultracompetent cells (Stratagene, catalog number: 200315)
- QIAquick gel extraction kit (QIAGEN, catalog number: 28704)
- Plasmid Mega/Giga purification kit (QIAGEN, catalog number: 12781)
- Retrovirus production and titration
- Phoenix–amphotropic retrovirus producing cells (Phoenix-AMPHO ATCC® CRL-3213TM)
- Dulbecco’s modified eagle medium (DMEM, high glucose) (Gibco, Life Technologies, catalog number: 11965092)
- Fetal bovine serum (Gibco, catalog number: 16000044)
- 200 mM L-Glutamine (Gibco, catalog number: 25030081)
- LipofectamineTM 2000 (Invitrogen, catalog number: 16680190)
- 0.45 μM SFCA membrane syringe filter (Corning Incorporated, catalog number: 443220)
- NIH3T3 mouse embryo fibroblast cells (ATCC, catalog number: CRL-1658)
- Transduction of viral cDNA library
- Retroviral cDNA library supernatant
- Polybrene (Hexadimethrine bromide, Sigma-Aldrich, catalog number: H9268)
- Cell culture
- Pipette tips and sterile serological pipettes
- 100-mm tissue culture-treated Petri-dish (BD Biosciences, catalog number: 353003)
- Tissue culture treated multiple well plates, 48-well (BD Bioscience, catalog number: 353078)
- Falcon tubes, polypropylene, 50-ml (BD Biosciences, catalog number: 352095)
- Falcon tubes, polypropylene, 15-ml (BD Biosciences, catalog number: 352096)
- Finely flame-drawn Pasteur pipettes
Fine Pasteur pipettes were made from commercial Pasteur pipettes (VWR, catalog number: 14673-043). The tip of pipette was heated several seconds on the flame of Bunsen burner, rapidly pulled out of flame using forceps, and cut the tip of pipette to obtain a suitable diameter. - (Optional) HamiltonTM Micro-syringe
Caution: Flame-drawn pipettes are very sharp. Handle with care. The pipette should be autoclaved and dried before use. - HMT3522 T4-2 cells (or any epithelial cancer cell line that can be ‘reverted’ to a phenotypically normal phenotype)
- Dulbecco’s modified Eagle’s medium (DMEM/F12, Thermo Fisher Scientific, catalog number: 12400-024)
- Insulin (250 ng/ml, Sigma-Aldrich, catalog number: I6634)
- Transferrin (10 μg/ml, Sigma-Aldrich, catalog number: T2252)
- Sodium Selenite (2.6 ng/ml, BD Bioscience, catalog number: 354201)
- Estradiol (10-10 M, Sigma-Aldrich, catalog number: E2758)
- Hydrocortisone (1.4 x 10-6 M, Sigma-Aldrich, catalog number: H0888)
- Prolactin (5 μg/ml, Sigma-Aldrich, catalog number: L6520)
- 0.25% (wt/vol) Trypsin (Thermo Fisher Scientific, catalog number: 25200-056)
- Soybean trypsin inhibitor (10 mg/ml, Sigma-Aldrich, catalog number: T6522)
- Na2HPO4 (Sigma-Aldrich, catalog number: S9390)
- KH2PO4 (J.T. Baker Inc., catalog number: 3246-01)
- NaCl (Sigma-Aldrich, catalog number: S5886)
- KCl (Sigma-Aldrich, catalog number: P5405)
- H14 Complete medium (without Epidermal Growth Factor) (see Recipes)
- 1x PBS (see Recipes)
- Phenotypic reversion assay on 3D lrECM culture
- LDEV-free growth factor reduced basement membrane matrix (MatrigelTM, Corning, catalog number: 354230; CultrexTM, Trevigen, catalog number: 3433-001-01)
- G418 Sulfate (Thermo Fisher Scientific, catalog number: 10131027) to select stably transduced cells
- Signaling pathway inhibitors (EGFR-TKI AG1478, EMD Millipore, catalog number: 658652; LY294002, Biomol, catalog number: L7751; Lapatinib or Gefitinib were kind gifts from Catherine Park at UCSF, but they are also commercially available at Sigma-Aldrich)
- Validation of selected clones
- Dispase II (neutral protease, grade II) (Sigma-Aldrich, catalog number: 4942078001)
- DNeasy® tissue kit (QIAGEN, catalog number: 69504)
- Primers to flanking sequences in the cloning vector
- LA Taq DNA Polymerase (A high fidelity Polymerase Chain Reaction (PCR) enzyme for long range) (Takara Bio, catalog number: RR002)
- pGEM®-T Easy cloning vector (Promega, catalog number: A1360)
Equipment
- Pipettes
- Water-Jacketed CO2 incubator (Thermo Scientific Forma, model: FormaTM Series II 3120)
- Centrifuge (Eppendorf, model: 5804)
- Inverted phase microscope (Nikon, model: Eclipse TS100)
- BioSafety cabinet
- Gene Pulse® electroporation cuvettes (Bio-Rad, catalog number: 1652089)
- Electroporation system (Bio-Rad Gene Pulser® II electroporation system, catalog numbers: 1652105-1652110)
- FluorchemTM 8900 gel imaging system (Alpha Innotech, model: 8900)
- PCR thermocycler
- (Optional) Micromanipulator (Burleigh, model: PCS5000)
Procedure
文章信息
版权信息
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Lee, S. and Bissell, M. J. (2018). A Functionally Robust Phenotypic Screen that Identifies Drug Resistance-associated Genes Using 3D Cell Culture. Bio-protocol 8(22): e3083. DOI: 10.21769/BioProtoc.3083.
分类
癌症生物学 > 癌症生物化学 > 耐药性
发育生物学 > 形态建成 > 器官形成
细胞生物学 > 细胞分离和培养 > 3D细胞培养
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link