- EN - English
- CN - 中文
Electroporation of Labeled Antibodies to Visualize Endogenous Proteins and Posttranslational Modifications in Living Metazoan Cell Types
利用标记抗体电穿孔来可视化研究后生动物活细胞中内源性蛋白和翻译后修饰
发布: 2018年11月05日第8卷第21期 DOI: 10.21769/BioProtoc.3069 浏览次数: 6243
评审: David CisnerosYi CuiTrinadh Venkata Satish Tammana
Abstract
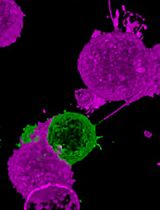
The spatiotemporal localization of different intracellular factors in real-time and their detection in live cells are important parameters to understand dynamic protein-based processes. Therefore, there is a demand to perform live-cell imaging and to measure endogenous protein dynamics in single cells. However, fluorescent labeling of endogenous protein in living cells without overexpression of fusion proteins or genetic tagging has not been routinely possible. Here we describe a versatile antibody-based imaging approach (VANIMA) to be able to precisely locate and track endogenous proteins in living cells. The labeling is achieved by the efficient and harmless delivery of fluorescent dye-conjugated antibodies or antibody fragments (Fabs) into living cells and the specific binding of these antibodies to the target protein inside of the cell. Our protocol describes step by step the procedure from testing of the suitability of the desired antibody, over the digestion of the antibody to Fabs until the labeling and the delivery by electroporation of the antibody or Fab into the cells. VANIMA can be adapted to any monoclonal antibody, self-produced or commercial, and many different metazoan cell lines. Additionally, our method is simple to implement and can be used not only to visualize and track endogenous factors, but also to specifically label posttranslational modifications, which cannot be achieved by any other labeling technique so far.
Keywords: Antibodies (抗体)Background
The fluorescent labeling of proteins to follow in real time their spatiotemporal localization in living cells was mainly achieved until now by using transgenic or overexpression-based approaches. However, the labeling of specific endogenous proteins or even posttranslational modifications in living cells is not yet routinely possible. Imaging of cellular structures and processes is typically performed by either immunofluorescence (IF) labeling on fixed cells or by exogenously overexpressing fluorescent fusion proteins in living cells. Although these well-established techniques showed to be very powerful to locate or follow proteins inside the cells, they inherit also some important drawbacks. In IF, the cells need to be chemically fixed and permeabilized to be able to incubate them with specific primary and secondary antibodies. Despite many variables and potential artifacts (Schnell et al., 2012; Teves et al., 2016) like fixation-related protein denaturation or permeabilization efficiency, IF is still often used to visualize target proteins in fixed cells or tissues. Otherwise, imaging of proteins in living cells is mainly achieved through the exogenously expression of fluorescent fusion proteins (Ellenberg et al., 1999; Betzig et al., 2006; Schneider and Hackenberger, 2017) or by knock-in of a fluorescent tag into the endogenous locus using the CRISPR/Cas9 technology (Ratz et al., 2015). Although fluorescent fusion proteins have been proven to be very powerful, they often do not behave as their endogenous counterparts due to their increased levels when exogenously overexpressed (Burgess et al., 2012). On the other hand, endogenous fusion proteins containing knocked-in tags are difficult to obtain as knock-in efficiencies are often very low. Consequently, there is a need for new and easy to implement imaging approaches to visualize endogenous target proteins in single living cells. Previous studies and methods, like FabLEM or the expression of mintbodies, showed that intracellular labeling of proteins with fluorescently labeled antibody fragments can give new insights into the dynamics of histone modifications (Hayashi-Takanaka et al., 2009; Hayashi-Takanaka et al., 2011; Sato et al., 2013). However, these techniques suffer from lower delivery efficiencies into living cells, or potential poor solubility of the intracellular expressed mintbodies. Recently, another method achieved fluorescent labeling of endogenous proteins by using a bacterial toxin called streptolysin O, which creates pores in the membrane of cells and allows for the delivery of fluorescent probes into living cells (Teng et al., 2016). However, this method requires additional steps to be able to reseal the membrane after treatment which can be quite harmful for the cells and can decrease cell viability. In contrast, our versatile antibody-based imaging approach (VANIMA) uses fluorescent dye-conjugated antibodies or Fabs, which are delivered into the cells by electroporation (Freund et al., 2013; Brees and Fransen, 2014). The antibody labeling reaction is highly efficient and can result in up to 5-7 fluorescent dyes per molecule of antibody depending on the antibody and the labeling kit used. The transduction of the antibodies has a very high delivery efficiency and viability of the cells is above 90% in human cancer cell lines such as U2OS. Afterwards, the transduced antibodies will bind to the endogenous target protein inside the cell and for nuclear targets they will be transported with the target protein into the nucleus (piggyback mechanism). Otherwise, for faster delivery into the nucleus of the cells, the antibodies can be digested to produce Fabs which can freely diffuse into the nucleus to find and bind their target. Thus, even proteins with posttranslational modifications in the nucleus can be visualized specifically using fluorescently-labeled Fabs against the target. Considering that there are several thousands of commercially-available antibodies that specifically recognize intracellular target proteins with high affinity, VANIMA can be used to uncover the dynamical behavior of a plethora of targets in living cells (Conic et al., 2018). Besides nuclear targets, the antibodies could also be used to label and image cytoplasmic structures/proteins. However, it is important to note that only proteins that are either directly accessible for the antibodies/Fabs or that can be reached through the piggyback mechanism can be labeled using this technique. We were already able to label α-tubulin in the cytoplasm but other accessible targets like the mitochondrial membrane or cytoplasmic vesicles could also be tested for labeling with VANIMA. However, specific labelling of cytoplasmic targets would only be possible if the target molecules are highly expressed. If their abundance in the cell is below the one of the introduced antibodies, a large fraction of the antibodies does not bind and will generate background staining. Additionally, the method is easy to implement in any laboratory and can also be used to perform multicolor imaging with different targets just by labeling two different antibodies with different dyes or by combining it with an already established endogenous knock-in clone. Finally, VANIMA can also be used with identified inhibiting antibodies to disrupt protein functions inside living cells.
Materials and Reagents
- 15 ml conical Falcon tubes (Corning, Falcon, catalog number: 352095)
- 1.5 ml Eppendorf tubes (Sigma-Aldrich, Eppendorf, catalog number: Z66505-100EA)
- 0.5 ml Eppendorf tubes (Sigma-Aldrich, Eppendorf, catalog number: Z666491-100EA)
- Falcon 12-well clear flat bottom cell culture plate (Corning, catalog number: 351143)
- µ-slide 8-well glass bottom: No. 1.5H (170 µm +/- 5 µm) (Ibidi, catalog number: 80827)
- 18 mm high precision cover glasses (Marienfeld, catalog number: 117580)
- Microscope slides ground edges plain (VWR, catalog number: 631-1552)
- Poly-Prep chromatography column (Bio-Rad, catalog number: 731-1550)
- DiaEasy dialyzer (3 ml) MWCO 6-8 kDa (Biovision, catalog number: K1013-25)
- DiaEasy dialyzer (800 µl) MWCO 6-8 kDa (Biovision, catalog number: K1019-25)
- Amicon Ultra-4 centrifugal filter units 10 kDa (Merck-Millipore, catalog number: UFC801024)
- Amicon Ultra-0.5 centrifugal filter units 10 kDa (Merck-Millipore, catalog number: UFC501096)
- CountessTM cell counting chamber slides (Thermo Fisher, catalog number: C10312)
- Sterile individually packaged 5 ml pipettes (Sigma-Aldrich, catalog number: SIAL1487)
- Sterile individually packaged 10 ml pipettes (Sigma-Aldrich, catalog number: SIAL1488)
- U2OS osteosarcoma cells [American Type Culture Collection (ATCC, catalog number: HTB-96)]
- Neon transfection 10 µl kit (including the Neon 10 µl tips) (Invitrogen, catalog number: MPK1096)
- AlexaFluor-488 antibody labeling kit (Invitrogen, catalog number: A20181)
- Dulbecco's Modified Eagle Medium (DMEM) (Thermo Scientific, Gibco, catalog number: 10567-014)
- Heat-inactivated fetal calf serum (FCS) (Gibco, catalog number: 15750-037)
- Gentamicin (Gibco, catalog number: 15750-037)
- 16% Paraformaldehyde (16% PFA) (Electron Microscopy Sciences, catalog number: 50-980-487)
- Phosphate Buffered Saline (PBS) (GE Healthcare, catalog number: SH30013.03)
- Triton X-100 (Sigma-Aldrich, catalog number: X100-100ML)
- Vectashield antifade mounting medium with DAPI (Vector-Laboratories, catalog number: H-1200-10)
- Alexa Fluor 488 goat-anti-mouse IgG (life technologies, catalog number: A11001)
- Protein G Sepharose FastFlow (GE Healthcare, catalog number: GE17-0618-01)
- Protein A Sepharose FastFlow (GE Healthcare, catalog number: GE17-5280-01)
- Papain-coated magnetic beads (Spherotech, catalog number: PAPM-40-2)
- Tris-HCl (Sigma-Aldrich, catalog number: 10812846001)
- Glycine (Sigma-Aldrich, catalog number: G8898)
- Sodium bicarbonate (Sigma-Aldrich, catalog number: S5761-1KG)
- Tris(2-carboxyethyl) phosphine hydrochloride (TCEP) (Sigma-Aldrich, catalog number: C4706)
- Sodium dodecyl sulfate (SDS) (Euromedex, catalog number: EU0660)
- Acrylamide/Bis-acrylamide 40% solution (Euromedex, catalog number: EU0077-B)
- Tetramethylethylendiamin (TEMED) (Serva, catalog number: 35930.01)
- Ammonium persulfate (APS) (Sigma-Aldrich, catalog number: A3678)
- Trypsin (Sigma-Aldrich, catalog number: T4799)
- U2OS growth medium (see Recipes)
- 4% Paraformaldehyde (4% PFA) (see Recipes)
- 10x Phosphate Buffered Saline (10x PBS) (see Recipes)
- Triton X-100 solutions (see Recipes)
- 10% Triton X-100
- 0.1% Triton X-100
- 0.02% Triton X-100
- 1 M Tris-HCl pH 8.2 (see Recipes)
- 0.1 M glycine-HCl pH 2.7 (see Recipes)
- Sodium bicarbonate buffers (see Recipes)
- 1 M sodium bicarbonate pH 8.2
- 0.1 M sodium bicarbonate
- 2.5% Trypsin (see Recipes)
Equipment
- Pipetman P2 pipette (Gilson, catalog number: F144801)
- Pipetman P20 pipette (Gilson, catalog number: F123600)
- Pipetman P200 pipette (Gilson, catalog number: F123601)
- Pipetman P1000 pipette (Gilson, catalog number: F123602)
- Jewelers forceps, Dumont No. 5 (Sigma-Aldrich, Dumont, catalog number: 6521)
- Pipette boy (Corning, Falcon, catalog number: 357469)
- Water bath (Julabo, model: ED (v.2))
- Magnetic tube rack (Diagenode, catalog number: B04000001)
- Fume hood Hera Safe KS (Thermo Scientific, catalog number: 51023175)
- Cell culture incubator with CO2 supply (Sanyo, catalog number: MCO-19AIC)
- SP8UV confocal microscope (Leica)
- Eppendorf centrifuge 5804 R (Eppendorf, model: 5804 R, catalog number: 805000620)
- Beckman Coulter Allegra centrifuge (Beckman, catalog number: 21R)
- NanoDrop 2000 spectrophotometer (Thermo Scientific, model: NanoDropTM 2000, catalog number: ND2000)
- Countess Cell Counter (Thermo-Fisher, catalog number: AMQAX1000)
- Neon Transfection System (Invitrogen, catalog number: MPK5000S)
Software
- Fiji/Image J (https://fiji.sc/)
Procedure
文章信息
版权信息
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Conic, S., Desplancq, D., Tora, L. and Weiss, E. (2018). Electroporation of Labeled Antibodies to Visualize Endogenous Proteins and Posttranslational Modifications in Living Metazoan Cell Types. Bio-protocol 8(21): e3069. DOI: 10.21769/BioProtoc.3069.
- Conic, S., Desplancq, D., Ferrand, A., Fischer, V., Heyer, V., Reina San Martin, B., Pontabry, J., Oulad-Abdelghani, M., Babu, N. K., Wright, G. D., Molina, N., Weiss, E. and Tora, L. (2018). Imaging of native transcription factors and histone phosphorylation at high resolution in live cells. J Cell Biol 217(4): 1537-1552.
分类
细胞生物学 > 基于细胞的分析方法 > 转运
免疫学 > 抗体分析 > 抗体检测
细胞生物学 > 细胞成像 > 活细胞成像
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link