- EN - English
- CN - 中文
Selective Isolation of Retroviruses from Extracellular Vesicles by Intact Virion Immunoprecipitation
通过完整病毒免疫沉淀技术从细胞外囊泡选择性分离逆转录病毒
发布: 2018年09月05日第8卷第17期 DOI: 10.21769/BioProtoc.3005 浏览次数: 12156
评审: Vamseedhar RayaproluSmita NairRajesh Thippeshappa
Abstract
There exists a wide variety of techniques to isolate and purify viral particles from cell culture supernatants. However, these techniques vary greatly in ease of use, purity, yield and impact on viral structural integrity. Most importantly, it is becoming evident that secreted extracellular vesicles (EVs) co-purify with retroviruses using nearly all purification methods due to nearly indistinguishable biophysical characteristics such as size, buoyant density and nucleic acid content. Recently, our group has illustrated a means of isolating intact and highly enriched retroviral virions from EV-containing cell supernatants using an immunoprecipitation approach targeting the viral envelope glycoprotein of the Moloney Murine Leukemia Virus (Renner et al., 2018). This technique, that we call intact virion immunoprecipitation (IVIP), enabled us to characterize the accessibility of epitopes on the surface of these retroviruses and assess the orientation of the virus-encoded integral membrane protein Glycogag (gPr80) in the viral envelope. Proper implementation of this protocol enables fast, simple and reproducible preparations of intact and highly purified retroviral particles devoid of detectable EV contaminants.
Keywords: Retrovirus purification (逆转录病毒纯化)Background
Widely used approaches for isolating retroviruses, such as the Human Immunodeficiency Virus (HIV) and Murine Leukemia Virus (MLV), include precipitation, chromatography, ultrafiltration, ultracentrifugation, as well as various other means of particle separation (Reviewed in Nestola et al., 2015). While each technique has its specific advantages, drawbacks and limitations, a common concern for all methods is the co-purification of cell secreted extracellular vesicles (EVs).
EVs constitute a heterogeneous population of membrane-derived vesicles secreted by all cell types (Yanez-Mo et al., 2015). There are strikingly similar biophysical and biochemical characteristics between retroviruses and EVs (Table 1), especially with the small 50-150 nm vesicles secreted through the endosomal pathway, better known as exosomes (Reviewed by Nolte-'t Hoen et al., 2016). Some retroviruses, such as HIV and MLVs, can also share with exosomes the pathways of biogenesis and egress through the endocytic system (Orenstein et al., 1988; Raposo et al., 2002; Houzet et al., 2006; Sandrin and Cosset, 2006; Akers et al., 2013; Madison and Okeoma, 2015; Martin et al., 2016; Nolte-'t Hoen et al., 2016). This imparts inherent biochemical composition similarities between the two types of particles, which extend to their cargo (e.g., proteins, mRNAs, miRNAs) and host-derived surface membrane proteins and antigens (e.g., CD9, CD63, CD81), which inevitably increases difficulties in telling them apart (Eckwahl et al., 2016; Nolte-'t Hoen et al., 2016; Telesnitsky and Wolin, 2016). Given such similarities, selective isolation of retroviruses requires a unique identifying marker to confidently discriminate them from exosomes and EVs in general.
Table 1. Retroviruses and EVs are nearly indistinguishable by their biochemical and biophysical characteristics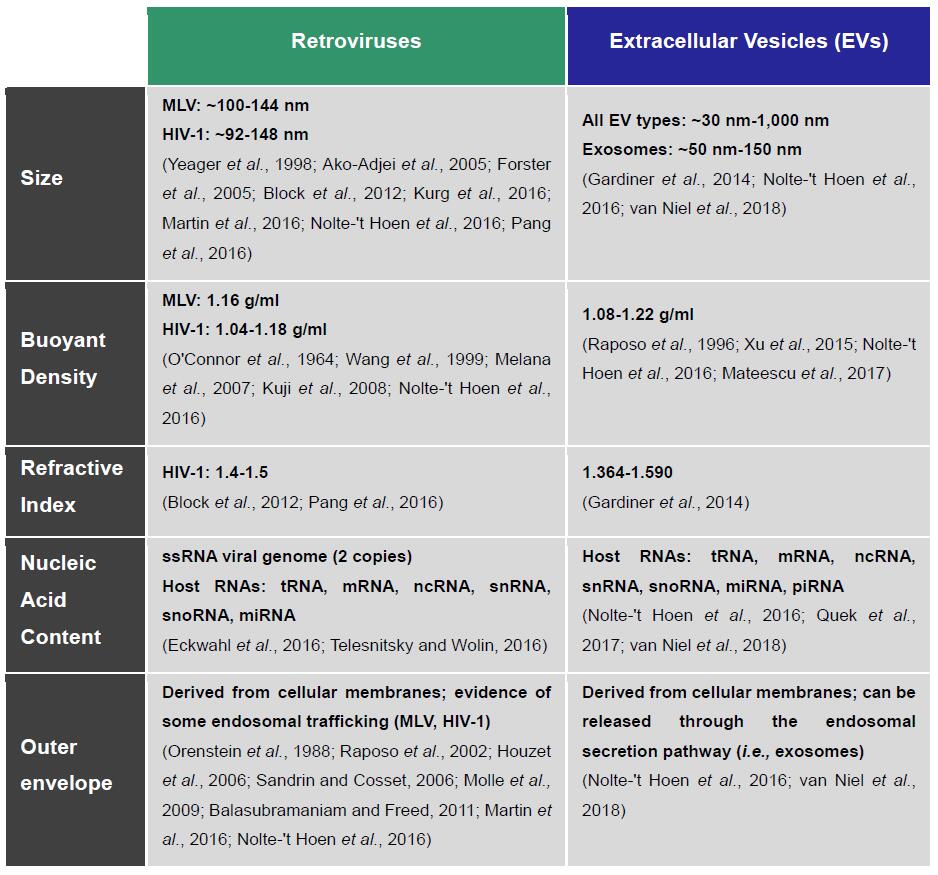
Nanoscale flow cytometry (NFC), also called flow virometry or NanoFACS, is an optimization of flow cytometry techniques, sample preparations and hardware for the analysis of particles smaller than 200 nm, which is the average detection limit of most commercial flow cytometers (Tang et al., 2016 and 2017; Lippe, 2018). This technology is especially useful for the immune phenotypic profiling of markers on the surface of viruses and EVs. By using this approach, we previously determined that a fluorescently tagged viral envelope glycoprotein (Env-eGFP) of the Moloney MLV (M-MLV) was almost exclusively expressed on the surface of these virus particles, and thereby constituted a very reliable selection marker (Figure 1) (Tang et al., 2017).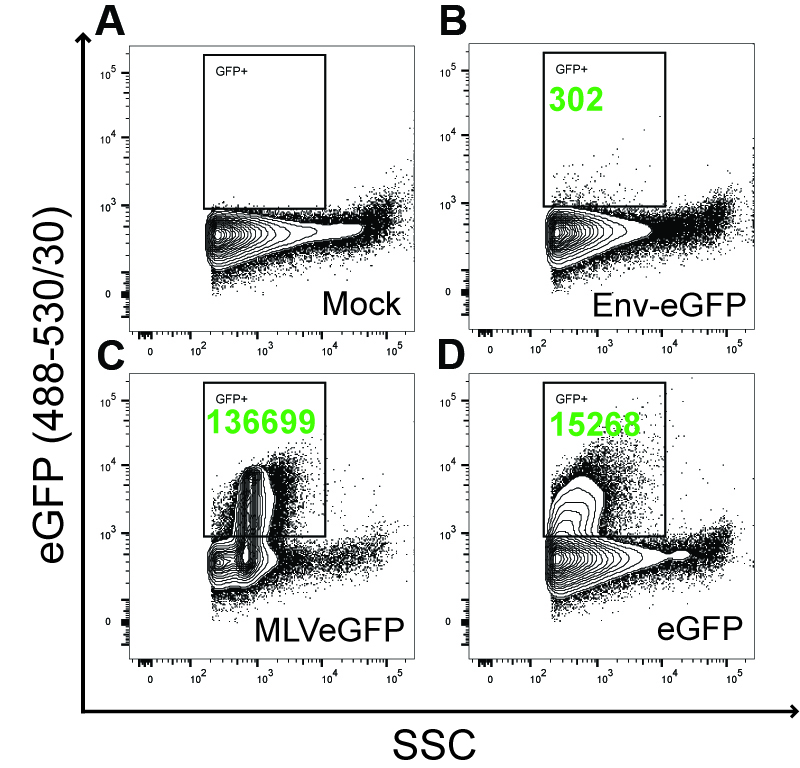
Figure 1. Env-eGFP represents a selection marker for identifying retroviruses by nanoscale flow cytometry. This figure has been adapted from Tang et al. (2017). 293T cells were mock transfected with an empty plasmid (A), with Env-eGFP (B) or eGFP (D) expression plasmids, or with the Env-eGFP expressing M-MLV viral plasmid (C). The M-MLV used in this study contained an eGFP reporter inserted into the proline-rich region of the extracellular domain of the envelope glycoprotein (Sliva et al., 2004). Supernatants were 450 nm-syringe filtered prior to NFC analysis. Transfection efficiency was monitored by eGFP expression in the transfected cells and was similar in each relevant condition (data not shown). For NFC analysis, particles were detected by triggering off of side-scattered light (SSC). A square gate was set above background in the Mock sample where eGFP+ events are expected (A). Side and top boundaries of this gate were determined by the limits of the eGFP+ events in the MLVeGFP sample (C). Numbers in green represent eGFP+ particles detected and enumerated in the gate during a fixed acquisition time window, which was the same for all samples analyzed. For NFC analysis, SSC is more sensitive than forward scattered (FSC) light to detect particles smaller than 200nm on our instrument (Tang et al., 2016). The results show that, in our system, the membrane-expressed Env-eGFP does not substantially associate with EVs (B). However, cytosolic eGFP is incorporated as cargo inside EVs (D). Env-eGFP is highly enriched only on the surface of viruses (C).
The protocol described here was specifically developed to study an enigmatic virus-encoded integral membrane protein called Glycogag (or gPr80) inserted in the envelope of M-MLV (Pillemer et al., 1986; Fujisawa et al., 1997 and 2001; Rosales Gerpe et al., 2015; Renner et al., 2018). Our goal was to assess the incorporation and orientation of full-length gPr80 in the envelope of M-MLV. A major caveat to this particular study was the release of EVs by the infected cells that contaminated our virus samples. This proved to be especially problematic, as we found that the gPr80 protein associated with both EVs and virions (Renner et al., 2018). But given that Env-eGFP was highly enriched on the surface of M-MLV virions and poorly incorporated on EVs (Figure 1) (Tang et al., 2017), we thus developed an intact virion immunoprecipitation (IVIP) assay designed to specifically isolate structurally intact viral particles expressing Env-eGFP on their surface. Using this approach, we successfully identified the orientation of gPr80 as a Type-I integral membrane protein on virions but as a Type-II integral membrane protein on EVs that are devoid of Env-eGFP (Renner et al., 2018). In conclusion, IVIP has the ability to selectively isolate and discriminate retroviruses from EVs with minimal physical manipulation and without compromising the structural integrity of either particle type.
Materials and Reagents
- μ-Columns (Miltenyi Biotec, catalog number: 130-042-701 )
- Microcentrifuge tubes (FroggaBio, catalog number: LMCT1.7B , or equivalent)
- Pasteur pipettes (Fisher Scientific, catalog number: 13-678-20A , or equivalent)
- PVDF membrane (Bio-Rad Laboratories, catalog number: 1620177 )
- Serological pipettes, 10 ml (Corning, catalog number: 4488 , or equivalent)
- Sterile 20 ml syringes with Luer-Lok (BD, catalog number: 302830 , or equivalent)
- Sterile 450 nm Luer-Lok syringe filters (Pall, catalog number: 4614 , or equivalent)
- Sterile 50 ml conical tubes (FroggaBio, catalog number: TB50-500 , or equivalent)
- Sterile pipette tips (Diamed, DIATEC, catalog numbers: DIATEC520-5376 , DIATEC520-5876 , DIATEC520-6501 , or equivalent)
- HEK 293T cells (ATCC, catalog number: CRL-3216 )
- R187 Hybridoma (ATCC, catalog number: CRL-1912 )
- μMACS GFP Isolation Kit (Miltenyi Biotec, catalog number: 130-091-125 )
- 10 cm culture dishes (Corning, catalog number: 430167 , or equivalent)
- 220 nm Steritop filters (Merck, catalog number: SCGPT10RE , or equivalent)
- Anti-eGFP (Takara Bio, Clontech, catalog number: 632381 )
- Anti-Flag, HRP conjugated (Sigma-Aldrich, catalog number: A8592-1MG )
- Anti-Mouse IgG, HRP conjugated (Cell Signaling Technology, catalog number: 7076S )
- Anti-Rabbit IgG, HRP conjugated (Abcam, catalog number: ab6721 )
- Anti-Rat IgG, HRP conjugated (Sigma-Aldrich, catalog number: AP183P )
- Anti-V5 (Merck, catalog number: AB3792 )
- Dulbecco's modified Eagle’s medium (DMEM) high glucose, with L-glutamine, sodium pyruvate and phenol red (WISENT, catalog number: 319-005-CL , or equivalent)
- Dynabeads M270-epoxy (Thermo Fisher Scientific, catalog number: 14321D )
- ECL Substrates, i.e.:
Clarity Western ECL Substrate (Bio-Rad Laboratories, catalog number: 1705060S , or equivalent)
ClarityMax Western ECL Substrate (Bio-Rad Laboratories, catalog number: 1705062S , or equivalent) - Fetal bovine serum (FBS) (Thermo Fisher Scientific, GibcoTM, catalog number: 12483020 , or equivalent)
- Glycine (Fisher Scientific, catalog number: BP381-5 )
- HCl (36.5-38%) (Fisher Scientific, catalog number: A144S-500 )
- Hybridoma-SFM (Thermo Fisher Scientific, GibcoTM, catalog number: 12045076 )
- KCl (Fisher Scientific, catalog number: BP366-500 )
- KH2PO4 (Fisher Scientific, catalog number: P285-500 )
- Methanol (VWR, catalog number: 56902-543 )
- Milli-Q Water
- Na2HPO4 (Fisher Scientific, catalog number: S393-3 )
- NaCl (Fisher Scientific, catalog number: BP358-10 )
- NaOH, 10N certified (Fisher Scientific, catalog number: SS255-1 )
- NuPAGETM 4-12% Bis-Tris Gel (Thermo Fisher Scientific, InvitrogenTM, catalog number: NP0335BOX )
- NuPAGETM MOPS SDS Running Buffer (Thermo Fisher Scientific, InvitrogenTM, catalog number: NP0001 )
- Penicillin-Streptomycin (GE Healthcare, catalog number: SV30010 , or equivalent)
- Polyethylenimine (PEI) (Polysciences, catalog number: 23966-1 , or equivalent)
- Sucrose (WISENT, catalog number: 800-081-LG , or equivalent)
- Tris Base (Fisher Scientific, catalog number: BP152-5 )
- TweenTM 20 (Fisher Scientific, catalog number: BP337-100 )
- PBS (10x) (see Recipes)
- 20% sucrose in PBS (see Recipes)
- Complete DMEM (see Recipes)
- Tris-Glycine Transfer Buffer (25x) (see Recipes)
Note: We used VWR as a distributor for Pall and Corning products.
Equipment
- μMACS Separator (Miltenyi Biotec, catalog number: 130-042-602 )
- 4 °C refrigerator
- Balance (Fisher Scientific, catalog number: 01-919-358, or equivalent)
Manufacturer: OHAUS, catalog number: 30100606/RM . - Biosafety cabinet (Thermo Fisher Scientific, catalog number: 1323TS , or equivalent)
- Digital Imager (GE Healthcare, model: ImageQuant LAS 4000, catalog number: 28955810 , or equivalent)
- Haemocytometer (Hausser Scientific, catalog number: 3100 , or equivalent)
- MACS MultiStand (Miltenyi Biotec, catalog number: 130-042-303 )
- Magnetic stand (Thermo Fisher Scientific, catalog number: 12321D )
- Microscope (Fisher Scientific, catalog number: LMI6PH2, or equivalent)
Manufacturer: Laxco, catalog number: LMI6PH2 . - Pipettes (Gilson, catalog number: F167700 , or equivalent)
- Refrigerated table-top centrifuge (Thermo Fisher Scientific, model: SorvallTM ST 40 , catalog number: 75004524, or equivalent)
- Rocking platform (Maxi Rotator) (Labline Instruments, model: Model 4631 , or equivalent)
- Tissue culture incubator, humidity, temperature and CO2 regulated (Thermo Fisher Scientific, catalog number: 3110 , or equivalent)
- Tube Revolver (Thermo Fisher Scientific, catalog number: 88881002 or equivalent)
- Type 70Ti Rotor (Beckman Coulter, catalog number: 337922 , or equivalent)
- Type 70Ti Tubes (Polycarbonate tubes and lids) (Beckman Coulter, catalog number: 355618 , or equivalent)
- Ultracentrifuge (Beckman Coulter, catalog number: 969347 , or equivalent)
Procedure
文章信息
版权信息
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Renner, T. M., Bélanger, K. and Langlois, M. (2018). Selective Isolation of Retroviruses from Extracellular Vesicles by Intact Virion Immunoprecipitation. Bio-protocol 8(17): e3005. DOI: 10.21769/BioProtoc.3005.
分类
微生物学 > 微生物生物化学 > 蛋白质 > 分离和纯化
微生物学 > 微生物生理学 > 膜性能
分子生物学 > 蛋白质 > 蛋白质-蛋白质相互作用
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link