- EN - English
- CN - 中文
Sleeping Beauty Transposon-based System for Rapid Generation of HBV-replicating Stable Cell Lines
使用基于睡美人转座子的系统快速生成HBV复制稳定细胞系
发布: 2018年07月05日第8卷第13期 DOI: 10.21769/BioProtoc.2908 浏览次数: 9700
评审: Yannick DebingVasudevan AchuthanAnonymous reviewer(s)
Abstract
The stable HBV-transfected cell lines, which based on stable integration of replication-competent HBV genome into hepatic cells, are widely used in basic research and antiviral drug evaluation against HBV. However, previous reported strategies to generate HBV-replicating cell lines, which primarily rely on random integration of exogenous DNA by plasmid transfection, are inefficient and time-consuming. We newly developed an all-in-one Sleeping Beauty transposon system (denoted pTSMP-HBV vector) for robust generation of stable HBV-replicating cell lines of different genotype. The pTSMP-HBV vector contains HBV 1.3-copy genome and dual selection markers (mCherry and puromycin resistance gene), allowing rapid enrichment of stably-transfected cells via red fluorescence-activated cell sorting and puromycin antibiotic selection. In this protocol, we described the detailed procedure for constructing the HBV-replicating stable cells and systematically evaluating HBV replication and viral protein expression profiles of these cells.
Keywords: HBV (HBV)Background
Chronic hepatitis B virus (HBV) infection is currently a major public health burden, affecting over 240 million individuals globally (Witt-Kehati et al., 2016). Patients with chronic HBV have an elevated risk of chronic active hepatitis, cirrhosis, or primary hepatocellular carcinoma (HCC) (Schweitzer et al., 2015). Current treatments with interferon-α or nucleoside analogs do not eradicate the virus, and their effects on clearing hepatitis B surface antigen (HBsAg) are limited (Lucifora and Protzer, 2016; Soriano et al., 2017). Therefore, there is an urgent need for the development of novel antiviral inhibitors (Nassal, 2015).
A cell culture model for evaluating the activity of new agents against HBV is an important tool for new drug development. The stable HBV-replicating cell lines, which carry replication-competent HBV genome stably integrated into the genome of human hepatoma cell lines (Huh7 and/or HepG2), are widely used to evaluate the effects of antiviral agents (Witt-Kehati et al., 2016). The stable HBV-producing human hepatoma cell lines (HepG2.2.15 and HepaAD38) integrated the D-genotype HBV genome, which are widely used in antiviral research (Chang et al., 1987; Ladner et al., 1997). However, stable HBV-producing cell lines of genotypes A, B, and C are not commonly used in the research field. Therefore, there is a need to develop cell lines of HBV genotypes A-C for drug development.
The Sleeping Beauty (SB) transposon system, derived from teleost fish sequences, is extremely effective at delivering DNA to vertebrate genomes, including those of humans (Structure of SB can be seen in Figure 1A). Sleeping Beauty transposition is a cut-and-paste process, during which the element ‘jumps’ from one DNA molecule to another (Figure 1B) (Ivics and Izsvak, 2011). Since its reconstruction in 1997 from the salmonid fish genome (Ivics et al., 1997), the SB system has been undergoing several modifications to improve its efficacy (Geurts et al., 2003; Baus et al., 2005; Score et al., 2006). The development of the hyperactive transposase SB100X has increased approximately 100-fold of efficiency compared with the first-generation transposase (Mátés et al., 2009), which is expected to facilitate widespread applications in functional genomics and gene therapy (Izsvak and Ivics, 2004).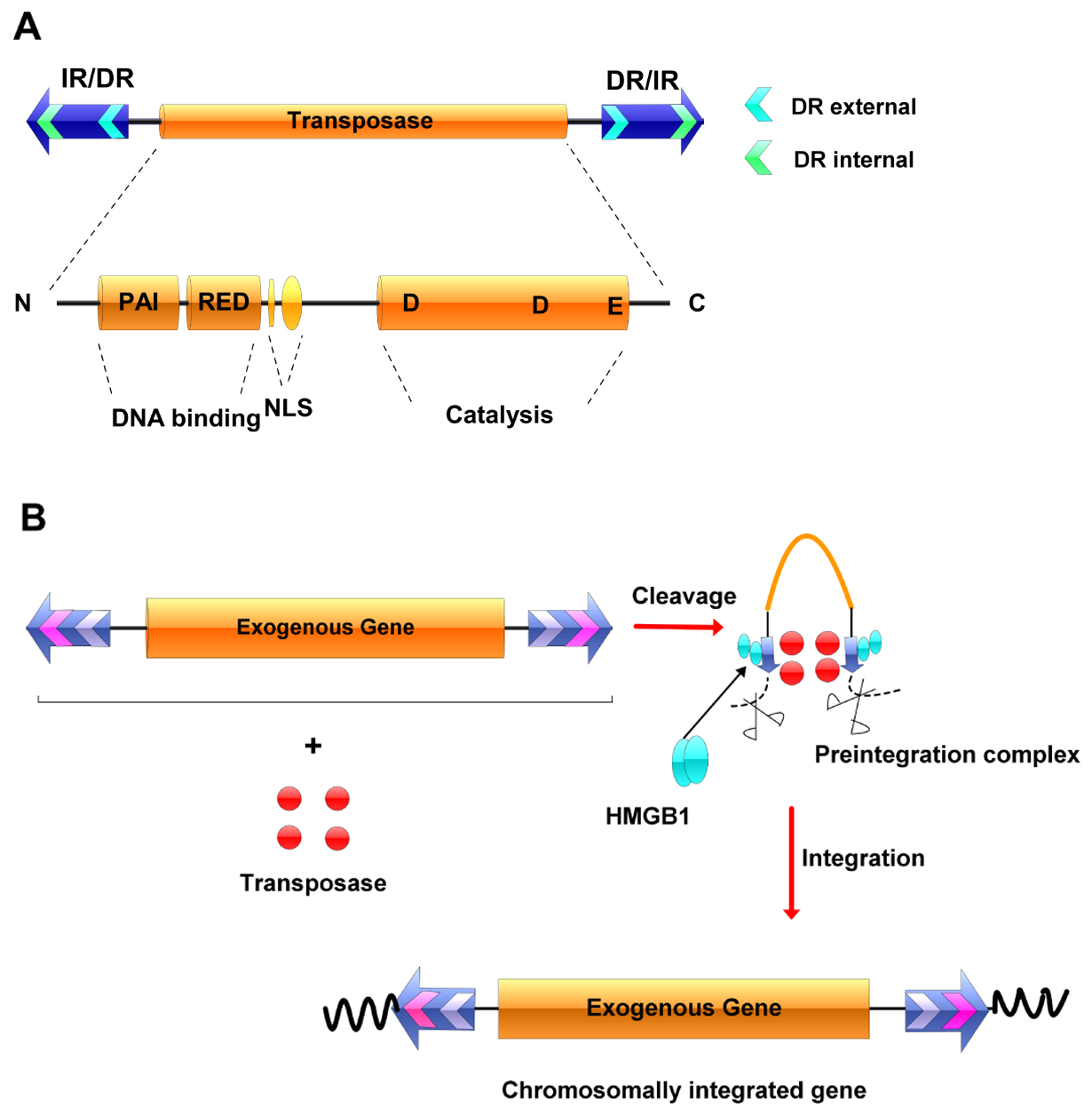
Figure 1. The Sleeping Beauty transposable element and its transposition. A. The Sleeping Beauty (SB) system. The transposase gene (yellow rectangle) is flanked by terminal inverted repeats (IR/DRs, blue arrows), each containing two binding sites for the transposase (small green arrows). The transposase consists of an N-terminal, DNA-binding domain (PAI and RED), a nuclear localization signal (NLS), a C-terminal and catalytic domain (DDE). B. Transposition. The transposase gene within the element can be replaced by a therapeutic gene, and the resultant transposon can be maintained in a simple plasmid vector. The transposase is supplied in trans. The transposase binds to its binding sites within the IR/DR repeats and, together with host factors such as HMGB1, forms a synaptic complex, in which the ends of the transposon are paired. The transposon is excised from the donor molecule and integrates into a new location.
Materials and Reagents
- Pipette tips, 10 μl (Haimen plastic, 20111088)
- Pipette tips, 200 μl (Corning, Axygen®, catalog number: T-200-Y-R )
- Pipette tips, 1 ml (Haimen plastic, 20111011)
- Cell culture plate (100 mm) (Corning, catalog number: 430167 )
- Cell culture plate (60 mm) (Thermo Fisher Scientific, catalog number: 150288 )
- 15 ml tube (Thermo Fisher Scientific, catalog number: 339651 )
- 70 μm cell strainer (Fisher Scientific, Fisherbrand, catalog number: 22-363-548 )
- Cell culture plate (6 well) (Corning, catalog number: 3516 )
- Cell culture plate (24 well) (Corning, catalog number: 3524 )
- Cell Imaging Plate, 24 well, glass bottom (Eppendorf, catalog number: 0030741021 )
- Nylon membranes (Roche Diagnostics, catalog number: 11417240001 )
- Human hepatoma HepG2 cells (Originally from the China Centre for Type Culture Collection, Wuhan, China)
- Minimum essential medium (MEM, powder) (Thermo Fisher Scientific, GibcoTM, catalog number: 41500-083 )
- Fetal bovine serum, Qualified, Australia Origin (Thermo Fisher Scientific, catalog number: 10099141 )
- X-tremeGENE HP DNA Transfection Reagent (Roche Diagnostics, catalog number: 06366236001 )
- Opti-MEM (Thermo Fisher Scientific, GibcoTM, catalog number: 31985070 )
- 0.25% Trypsin-EDTA (1x), Phenol Red (Thermo Fisher Scientific, GibcoTM, catalog number: 25200-114 )
- Puromycin (Thermo Fisher Scientific, InvitrogenTM, catalog number: A1113803 )
- Mouse anti-HBcAg (Innodx Biotechnology, catalog number: 2A7-21 [it's new anti-HBc mAb, available on request])
- Alexa Fluor® 488 Donkey Anti-Mouse IgG (H+L) (Thermo Fisher Scientific, catalog number: A-21202 )
- DAPI (Thermo Fisher Scientific, InvitrogenTM, catalog number: D1306 )
- Micrococcal nuclease (Takara Bio, catalog number: D2910 )
- Proteinase K (Takara Bio, catalog number: D9033 )
- Ethanol, C2H6O, AR (Xilong Scientific, catalog number: 1030029AR )
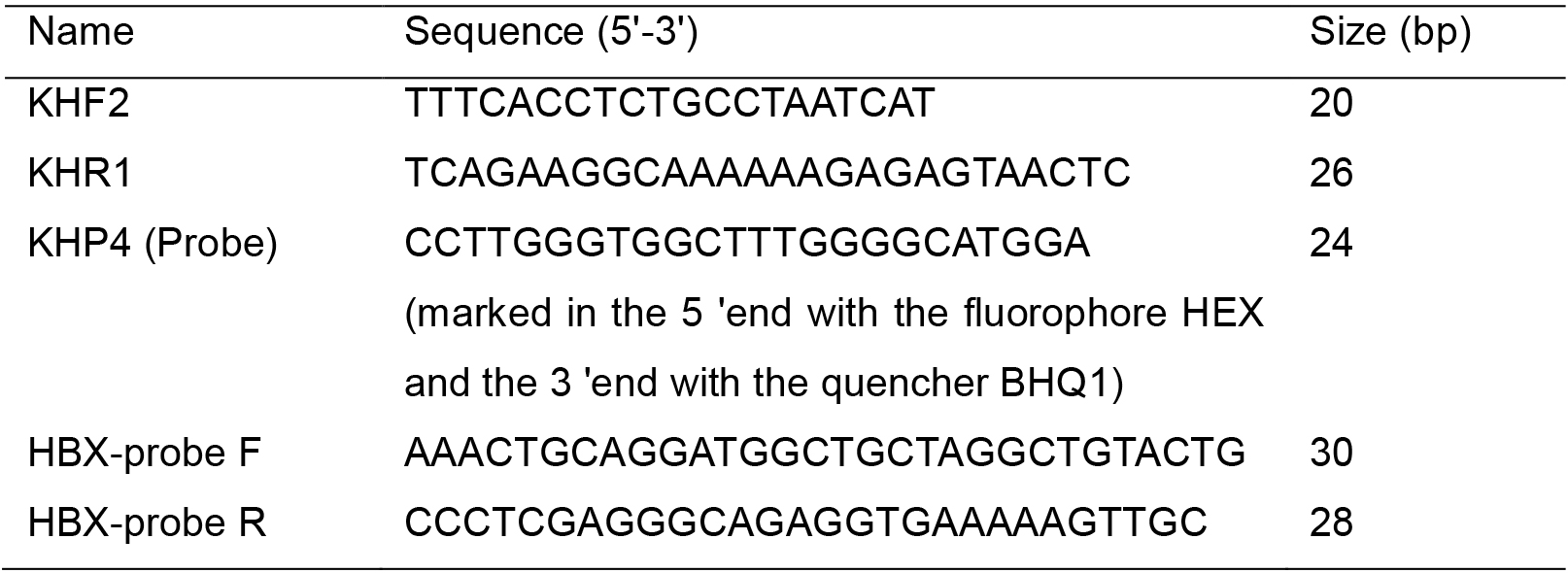
- Oligonucleotides
- dNTP Mixture 2.5 μM (Takara Bio, catalog number: D4030A )
- DIG Easy Hyb Granules (Roche Diagnostics, catalog number: 11796895001 )
- PrimeSTAR GXL DNA Polymerase (Takara Bio, catalog number: DR050A )
- Premix Ex TaqTM (Probe qPCR) (Takara Bio, catalog number: RR390A )
- Casein 10x blocking buffer (Sigma-Aldrich, catalog number: B6429-500ML )
- Anti-DIG (AP) antibody (Roche Diagnostics, catalog number: 11093274910 )
- CDP-Star AP substrate (Roche Diagnostics, catalog number: 12041677001 )
- Universal DNA Purification Kit (TIANGEN Biotech, catalog number: DP214-03 )
- Diagnostic kit for Hepatitis B surface antigen (CLEIA) (Wantai Biological Pharmacy, catalog number: HBV-1396 )
- Diagnostic kit for Hepatitis B e-antigen (ELISA) (Wantai Biological Pharmacy, catalog number: HBV-0396 )
- ED-11 (Innovax Biotechnology)
- Virus DNA/RNA Extraction kit (GenMag Biotechnology, catalog number: NA007 )
- Mycoplasma-free neonatal bovine serum (Tianhang Biotechnology, catalog number: 11011-8615 )
- Sodium chloride (NaCl, AR) (Xilong Scientific, catalog number: 1001012AR )
- Potassium chloride (KCl, cell culture) (Sigma-Aldrich, catalog number: P5405 )
- Hydrochloric acid (HCl, AR) (Xilong Scientific, catalog number: 1029013AR )
- Sodium hydroxide (NaOH, AR) (Xilong Scientific, catalog number: 1001037AR )
- Disodium hydrogen phosphate (Na2HPO4·12H2O, AR) (Xilong Scientific, catalog number: 1001067AR )
- Potassium dihydrogen phosphate (KH2PO4, AR) (Xilong Scientific, catalog number: 1002048AR500 )
- Sodium bicarbonate (NaHCO3, AR) (Xilong Scientific, catalog number: 44558 )
- Paraformaldehyde (Sigma-Aldrich, catalog number: 16005-1KG-R )
- Triton X-100 (AMRESCO, catalog number: 0694 )
- Bovine albumin (Low endotoxin) (ICPbio International, catalog number: ABRE-1KG )
- Tris base (SEEBIO BIOTECH, catalog number: 183995 )
- Tris-saturated phenol (Solarbio, catalog number: T0250 )
- Ethylenediaminetetraacetic acid (EDTA, AR) (Xilong Scientific)
- NONIDET® P-40 Substitute (AMRESCO, catalog number: M158-500ML )
- Calcium chloride (CaCl2, AR) (Xilong Scientific, catalog number: U1000566-500g )
- Sodium dodecyl sulfate (SDS) (Merck, catalog number: 428015 )
- Chloroform (AR) (Xilong Scientific, catalog number: 1039013AR500 )
- Isoamyl alcohol (AR) (Xilong Scientific, catalog number: U1001975-500ml )
- Maleic acid (Sigma-Aldrich, catalog number: M0375 )
- Tween-20 (BBI Solutions, catalog number: TB0560-500ml )
- Ethylenediaminetetraacetic acid disodium salt dihydrate (EDTA-Na2·2H2O, AR) (Xilong Scientific, catalog number: 100186 )
- Acetate (CH3COOH, AR) (Xilong Scientific, catalog number: 1029047AR )
- Sodium citrate (C6H5Na3O7, AR) (Xilong Scientific, catalog number: 1001059AR )
- Verson buffer (see Recipes)
- PBS buffer (see Recipes)
- 4% paraformaldehyde (see Recipes)
- 0.2% Triton X-100 (see Recipes)
- 3% BSA (see Recipes)
- NET buffer (see Recipes)
- 1.2 M CaCl2 (see Recipes)
- 0.5 M EDTA (see Recipes)
- 10% SDS (see Recipes)
- Phenol/chloroform/isoamyl alcohol (25:24:1) (see Recipes)
- Maleic acid buffer (see Recipes)
- Washing buffer (see Recipes)
- Detection buffer (see Recipes)
- 50x TAE buffer (see Recipes)
- Depurination buffer (see Recipes)
- Denaturation buffer (see Recipes)
- Neutralization buffer (see Recipes)
- 10x SSC (see Recipes)
- 2x SSC/0.1% SDS (see Recipes)
- 0.5x SSC/0.1% SDS (see Recipes)
Equipment
- Pipettes (Mettler-Toledo International, RAININ, model: Pipet-Lite )
- CO2 Incubator (Thermo Fisher Scientific, catalog number: 3111 )
- Centrifuge (Thermo Fisher Scientific, model: HeraeusTM PicoTM 17 )
- Sorvall refrigeration Centrifuge (Thermo Fisher Scientific, model: SorvallTM ST 16R )
- BD FACS Aria III (BD, model: FACSAriaTM III )
- High Content Screening System (PerkinElmer, model: Opera PhenixTM )
- Water bath (Grant Instruments, model: GD100 )
- UV cross-linking instrument (Shanghai SIGMA High-tech, model: SH4 )
- Bio-Rad vacuum blotter (Bio-Rad Laboratories, model: Model 785 )
- Multifunctional molecular hybridization oven (UVP, model: HM-4000 )
- ImageQuant LAS4000 mini (GE Healthcare, model: ImageQuant LAS 4000 mini )
- Plastic film sealing machine (Shanghai Mingwei, model: F-400 )
- Biomek NXP (Beckman Coulter, model: Biomek NXP )
- Microplate reader (Autobio, model: PHOmo )
- Orion II Microplate Lumimometer (Titertek-Berthold, model: Orion II )
- Electrophoresis apparatus (Bio-Rad Laboratories, catalog number: 1645050 )
- LightCycler® 96 (Roche Molecular Systems, model: LightCycler® 96 )
Procedure
文章信息
版权信息
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Zheng, J., Cao, J. and Yuan, Q. (2018). Sleeping Beauty Transposon-based System for Rapid Generation of HBV-replicating Stable Cell Lines. Bio-protocol 8(13): e2908. DOI: 10.21769/BioProtoc.2908.
分类
微生物学 > 微生物-宿主相互作用 > 体外实验模型 > 细胞系
微生物学 > 微生物-宿主相互作用 > 病毒
细胞生物学 > 细胞结构 > 染色体
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link