- EN - English
- CN - 中文
Extraction of Small Molecules from Fecal Samples and Testing of Their Activity on Microbial Physiology
从粪便样本中提取小分子并检测其对微生物的生理学活性
发布: 2018年04月20日第8卷第8期 DOI: 10.21769/BioProtoc.2808 浏览次数: 7985
评审: David CisnerosParul MehrotraAnonymous reviewer(s)

相关实验方案

番茄内生菌Streptomyces sp. VITGV100中吲哚类代谢产物的合成与提取分析
Veilumuthu Pattapulavar [...] John Godwin Christopher
2025年07月20日 2405 阅读
Abstract
The human body is colonized by vast communities of microbes, collectively known as microbiota, or microbiome. Although microbes colonize every surface of our bodies that is exposed to the external environment, the biggest collection of microbes colonizing humans and other mammals can be found in the gastrointestinal tract. Given the fact that the human gut is colonized by several hundred microbial species, our group hypothesized that the chemical diversity of this environment should be significant, and that many of the molecules present in that environment would have important signaling roles. Therefore, we devised a protocol to extract these molecules from human feces and test their signaling properties. Potentially bioactive extracts can be tested through addition to culture medium and analyses of bacterial growth and gene expression, among other properties. The protocol described herein provides an easy and rapid method for the extraction and testing of metabolites from fecal samples using Salmonella enterica as a model organism. This protocol can also be adapted to the extraction of small molecules from other matrices, such as cultured mammalian cells, tissues, body fluids, and axenic microbial cultures, and the resulting extracts can be tested against various microbial species.
Keywords: Metabolome (代谢组学)Background
Complex assemblages of microbes live in and on humans, colonizing every surface exposed to the external environment. These communities have received several denominations over the decades, including normal flora, microbiota, and, more recently, microbiome (Sekirov et al., 2010; Kashyap et al., 2017). In humans, these vast microbial communities colonize our skin, respiratory tract, genitals, gastrointestinal tract, and many other sites. By far, the most heavily colonized site is the gastrointestinal tract, where trillions of microbes, encompassing several hundred species, coexist peacefully with their hosts. Some of these species knowingly live in symbiotic associations with the human organism, where both parts benefit from the interactions. For others, the relationship may be purely commensal, where the parts coexist without causing any harm to each other, but without providing or obtaining any benefit (Sekirov et al., 2010; Kundu et al., 2017).
The human gastrointestinal microbiota presents significant diversity in their composition, and stands out as a complex environment where interactions between different microbes as well as between microbes and host cells constantly occur. Bacteria are known to produce a plethora of bioactive small molecules, such as antibiotics, bacteriocins, pigments, secondary metabolites, quorum sensing signals, and many others (Antunes and Ferreira, 2009; Antunes et al., 2010; Antunes et al., 2011a). In such a complex environment as the intestinal microbiome, it is almost imperative to consider the production and accumulation of such molecules. These small molecules may represent by-products of metabolic activities or signals with specific roles, and can be produced both by the host itself as well as the microbes living in that environment; in many cases these small molecules are the tools used by these organisms to interact. Using high-throughput mass spectrometry-based metabolomics, we have previously shown that thousands of small molecules can be found in the lumen of the mammalian intestinal tract, and that the gut microbiome is involved in the production of many of them (Antunes et al., 2011b). In order to ascertain the signaling potential of small molecules from the gut metabolome, we have also extracted these molecules and tested their ability to modulate growth and gene expression of an enteric pathogen. As shown by our previous results, Salmonella enterica serovar Typhimurium responds to these molecules, and modulates the expression of over one hundred genes in response to bioactive small molecules from human feces (Antunes et al., 2014). Interestingly, many of the genes regulated by the fecal extract are required for the pathogenesis of Salmonella, such as those involved in the invasion of non-phagocytic host cells. More recently, we were able to purify and identify small aromatic compounds as the culprits for the regulation of Salmonella genes by the human gut metabolome (Peixoto et al., 2017). Here, we describe in detail the methods used by our group to obtain small molecules from the human gut metabolome and test them against Salmonella for various biological activities. A workflow of the procedures described herein can be found in Figure 1. 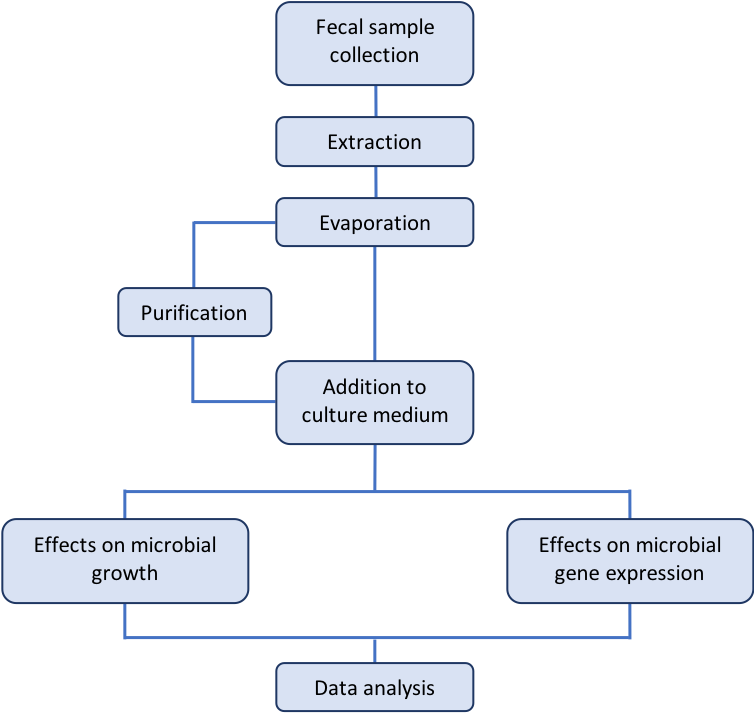
Figure 1. Workflow of the procedures described in this protocol
Materials and Reagents
- Polypropylene container (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 193A )
- Aluminum foil
- Tape
- Graduated glass pipettes (Fisher Scientific, catalog number: 13-678-25E )
- 2-ml Safe-Lock tubes (Eppendorf, catalog number: 0030120094 )
- Syringes (Descarpack, catalog number: 0324501 )
- Conical tubes (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 362694 )
- Axygen universal pipette tips (Corning, Axygen®, catalog number: T-200-C-L-R )
- Barrier tips (Fisher Scientific, catalog number: 02-707-430 )
- Syringe filter, 0.22-µm pore (KASVI, catalog number: K18-230 )
- Cuvettes (BRAND, catalog number: 759115 )
- Inoculation loop
- Borosilicate tubes, 16 x 100 mm (DWK Life Sciences, Kimble, catalog number: 73500-16100 )
- Bacterial culture (Salmonella enterica serovar Typhimurium SL1344)
- HPLC-grade ethyl acetate, ≥ 99.7% pure (Sigma-Aldrich, catalog number: 34858 )
- Nitrogen gas
- HPLC-grade methanol (Sigma-Aldrich, catalog number: 34860 )
- C18 cartridges containing 360 mg of sorbent (WATERS, catalog number: WAT051910 )
- Distilled water
- Phosphate-buffered saline (Sigma-Aldrich, catalog number: P5493-1L )
- RNeasy Mini Kit (QIAGEN, catalog number: 74106 )
- Agarose (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 17850 )
- QuantiTect Reverse Transcription Kit (QIAGEN, catalog number: 205311 )
- MinElute Reaction Cleanup Kit (QIAGEN, catalog number: 28204 )
- Primers (Integrated DNA Technologies)
- Power SYBR Green PCR Master Mix (Thermo Fisher Scientific, Applied BiosystemsTM, catalog number: 4367659 )
- Hydrochloric acid, 36.5-38% (Sigma-Aldrich, catalog number: H1758 )
- Sodium hydroxide, ≥ 98% pure (Sigma-Aldrich, catalog number: S8045 )
- Luria-Bertani (LB) Broth (BD, catalog number: 244620 )
- Bacteriological agar (Sigma-Aldrich, catalog number: A5306 )
- Streptomycin (Sigma-Aldrich, catalog number: S9137-25G )
- 1 N HCl (see Recipes)
- 1 N NaOH (see Recipes)
- LB broth or agar (see Recipes)
- LB with streptomycin (see Recipes)
Equipment
- Digital scale (Ohaus, model: SP202 )
- Glass bottle (Fisher Scientific, catalog number: FB800500 )
- Orbital shaker (BiomiXer, model: TS-2000A )
- Graduated glass cylinder (Laborglas, catalog number: 91376 )
- Glass boiling flask (Corning, PYREX®, catalog number: 4100-125 )
- Electronic pipette (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 9501 )
- Fume hood
- Rotary evaporator (Heidolph, catalog number: 560-01300-00 )
- 37 °C incubator
- Freezer, -20 °C
- P20 micropipette (Gilson, catalog number: F123600 )
- P200 micropipette (Gilson, catalog number: F123601 )
- P1000 micropipette (Gilson, catalog number: F123602 )
- Speed-Vac concentrator (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: SPD131DDA-115 )
- Vortex (Scientific Industries, model: Vortex-Genie 2, catalog number: SI-0236 )
- pH meter (Sigma-Aldrich, catalog number: MT 51302916 )
Manufacturer: Mettler-Toledo, catalog number: 51302916 . - Autoclave
- Visible range spectrophotometer
- Nucleic acid (UV) spectrophotometer
- Shaker (NOVATECNICA, model number: NT145 )
- Centrifuge for 96-well plates (Eppendorf, model: 5810 , catalog number: 5810000017)
- Real Time PCR Machine (Thermo Fisher Scientific, Applied BiosystemsTM, catalog number: 4376357 )
Software
- GraphPad Prism (GraphPad Software)
- StepOne Software v2.3 (Thermo Fisher Scientific)
Procedure
文章信息
版权信息
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Alves, E. S., Ferreira, R. B. R. and Antunes, L. C. M. (2018). Extraction of Small Molecules from Fecal Samples and Testing of Their Activity on Microbial Physiology. Bio-protocol 8(8): e2808. DOI: 10.21769/BioProtoc.2808.
分类
微生物学 > 微生物信号传导 > 群体感应
生物化学 > 其它化合物 > 小分子
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link












