- EN - English
- CN - 中文
Isolation of Commensal Escherichia coli Strains from Feces of Healthy Laboratory Mice or Rats
从健康实验小鼠或大鼠粪便中分离共生大肠埃希杆菌菌株
发布: 2018年03月20日第8卷第6期 DOI: 10.21769/BioProtoc.2780 浏览次数: 10898
评审: David CisnerosWolf Dieter RötherAnonymous reviewer(s)

相关实验方案
细菌病原体介导的宿主向溶酶体运输的抑制:基于荧光显微镜的DQ-Red BSA分析
Mădălina Mocăniță [...] Vanessa M. D'Costa
2024年03月05日 2893 阅读
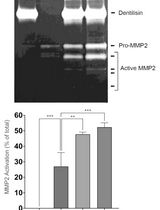
通过制备连续聚丙烯酰胺凝胶电泳和凝胶酶谱分析法纯化来自梭状龋齿螺旋体的天然Dentilisin复合物及其功能分析
Pachiyappan Kamarajan [...] Yvonne L. Kapila
2024年04月05日 2080 阅读
Abstract
The colonization abundance of commensal E. coli in the gastrointestinal tract of healthy laboratory mice and rats ranges from 104 to 106 CFU/g feces. Although very well characterized, the family that E. coli belongs to has a very homogeneous 16S rRNA gene sequence, making the identification from 16S rRNA sequencing difficult. This protocol provides a procedure of isolating and identifying commensal E. coli strains from a healthy laboratory mouse or rat feces. The method can be applied to isolate commensal E. coli from other laboratory rodent strains.
Keywords: Commensal (共生)Background
Escherichia coli is a Gram-negative, facultative anaerobe which constitutes only a minor fraction of the vertebrate gut microbiota, but plays a key role in microbial interaction, immune modulation and metabolic functionalities (Tenaillon et al., 2010). Being one of the best-characterized model microorganisms, commensal E. coli strains have been increasingly studied to unravel the mechanisms through which gut commensal microbes adapt to the unique niche and impact host physiology. However, the high homology among different strains raises difficulties in identification and characterization of commensal E. coli based on a 16S rRNA sequencing approach. Thanks to the development of next-generation sequencing techniques and large-scale analyses of whole genomes, we are able to identify commensal E. coli strains isolated from the gastrointestinal tract of different hosts according to the presence of virulence genes in the genome. In this protocol, we show an approach to isolate and identify commensal E. coli strains from a laboratory mouse or rat using selective culture media and whole genome sequencing. However, it should be noted that the presence of commensal E. coli in laboratory animals depends on the vendor and environmental conditions of the facility.
Materials and Reagents
- Gloves and masks (KCWW, Kimberly-Clark, catalog number: 52817 ; Cardinal Health, Insta-Gard, catalog number: AT7511-WE )
- 1.5 ml centrifuge tube (sterile) (Fisher Scientific, catalog number: 05-408-129 )
- Wide bore tips, 0-200 μl (Corning, Axygen®, catalog number: T-1005-WB-C )
- Thin-wall PCR tubes (Fisher Scientific, catalog number: 14-230-225 )
- Tips, 0.1-10 μl, 0.1-1 ml (sterile) (Fisher Scientific, catalog numbers: 02-707-474 ; 02-707-480 )
- Cell spreader (Fisher Scientific, catalog number: 08-100-11 )
- Petri dishes (100 x 15 mm) (Fisher Scientific, catalog number: FB0875713 )
- 15 ml conical sterile polypropylene centrifuge tubes (Thermo Fisher Scientific, NuncTM, catalog number: 339650 )
- Sterile 0.22 μm filter (Corning, catalog number: 431219 )
- 1 ml syringe (BD, catalog number: 309659 )
- A healthy NIH Swiss mouse (Harlan Laboratories Inc., Indianapolis, IN) and Sprague-Dawley (SD) rat (Charles River Canada, St. Constant, QC)
- 70% ethanol diluted from 100% ethanol (Commercial Alcohols, catalog number: P016EAAN )
- Agarose (Thermo Fisher Scientific, InvitrogenTM, catalog number: 16500500 )
- Tris-Acetate-EDTA (TAE) (50x stock) (Fisher Scientific, catalog number: BP13321 )
- SYBR Safe DNA gel stain (Thermo Fisher Scientific, InvitrogenTM, catalog number: S33102 )
- 6x DNA loading dye (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: R0611 )
- 1 kb Plus DNA ladder (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: SM1331 )
- GeneJET gel extraction and DNA cleanup micro kit (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: K0832 )
- PureLink genomic DNA mini kit (Thermo Fisher Scientific, InvitrogenTM, catalog number: K182001 )
- Nextera XT DNA library preparation kit (Illumina, catalog number: FC-131-1096 )
- Nextera XT DNA library preparation index kit (Illumina, catalog number: FC-131-1002 )
- Nextera XT DNA library preparation kit (Illumina, catalog number: FC-131-1096 )
- Nextera XT DNA library preparation index kit (Illumina, catalog number: FC-131-1002 )
- QubitTM 1x dsDNA HS assay kit (Thermo Fisher Scientific, InvitrogenTM, catalog number: Q33230 )
- PhiX control kit (Illumina, catalog number: FC-110-3001 )
- MiSeq reagent kit V3 (Illumina, catalog number: MS-102-3003 )
- Sodium phosphate dibasic (Na2HPO4) (Fisher Scientific, catalog number: S373-500 )
- Potassium phosphate monobasic (KH2PO4) (Fisher Scientific, catalog number: P285-500 )
- Sodium chloride (NaCl) (Fisher Scientific, catalog number: S271-500 )
- Potassium chloride (KCl) (Fisher Scientific, catalog number: P217-500 )
- Hydrochloric acid (HCl) (Fisher Scientific, catalog number: A144-500LB )
- MacConkey agar (BD, catalog number: 212123 )
- Luria-Bertani (LB) broth (Sigma-Aldrich, catalog number: L3022 )
- Glycerol (Fisher Scientific, catalog number: BP229-1 )
- DNA Taq polymerase with 50 mM MgCl2 (Thermo Fisher Scientific, InvitrogenTM, catalog number: 10342020 )
- Oligo primers 27F/1492R (Weisburg et al., 1991)
27F: 5’-AGAGTTTGATCMTGGCTCAG-3’
1492R: 5’-TACGGYTACCTTGTTACGACTT-3’ - Deoxynucleotide triphosphates (dNTPs) (Thermo Fisher Scientific, InvitrogenTM, catalog number: 10297-018 )
- PCR grade water (Thermo Fisher Scientific, InvitrogenTM, catalog number: AM9932 )
- Sodium hydroxide (NaOH) (Fisher Scientific, catalog number: SS266-1 )
- 1x phosphate buffered saline (PBS) (pH 7.4) (see Recipes)
- MacConkey agar (see Recipes)
- LB broth (see Recipes)
- Glycerol stock of bacterial isolates (see Recipes)
- PCR reaction mix (see Recipes)
- 0.1 N NaOH (see Recipes)
Equipment
- Forceps (sterilized by autoclave)
- Vortex (Fisher Scientific, catalog number: 02215365 )
- Pipettes [e.g., P1000 (Eppendorf, catalog number: 3120000062 ), P200 (Eppendorf, catalog number: 3120000054 ), P10 (Eppendorf, catalog number: 3120000020 )]
- Thermal cycler (Thermo Fisher Scientific, Applied Biosystems, model: GeneAMP PCR System 9700 )
- Microwave (RCA, catalog number: RMW733 )
- Gel electrophoresis system (Fisher Scientific, model: FB300 )
- UV transilluminator with photo documentation (Azure Biosystems, model: Azure c200 )
- Incubator (37 °C) (Thermo Fisher Scientific, Thermo Scientific, model: Model 370 )
- Shaking incubator (37 °C) (Eppendorf, New BrunswickTM, model: I26 )
- QubitTM 3.0 fluorometer (Thermo Fisher Scientific, InvitrogenTM, catalog number: Q33216 )
- QubitTM assay tubes (Thermo Fisher Scientific, InvitrogenTM, catalog number: Q32856 )
- Illumina MiSeq instrument (Illumina, model: MiSeqTM System, catalog number: SY-410-1003 )
- Autoclave (Beta Star Life Science Equipment, model: C2002BS )
- -80 °C freezer (Thermo Fisher Scientific, Thermo ScientificTM, model: FormaTM 900 Series , 989)
Procedure
文章信息
版权信息
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Ju, T. and Willing, B. P. (2018). Isolation of Commensal Escherichia coli Strains from Feces of Healthy Laboratory Mice or Rats. Bio-protocol 8(6): e2780. DOI: 10.21769/BioProtoc.2780.
分类
微生物学 > 微生物-宿主相互作用 > 细菌
微生物学 > 微生物细胞生物学 > 细胞分离和培养
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link