- EN - English
- CN - 中文
Detection of Protein Interactions in the Cytoplasm and Periplasm of Escherichia coli by Förster Resonance Energy Transfer
采用Förster共振能量转移法检测大肠埃希杆菌细胞质和周质中蛋白质的相互作用
发布: 2018年01月20日第8卷第2期 DOI: 10.21769/BioProtoc.2697 浏览次数: 14444
评审: Valentine V TrotterEric DurandVikash Verma

相关实验方案
蛋白质pull-down免疫共沉淀实验分析病毒DNA结合蛋白质之间的直接相互作用
Ana Lechuga [...] Modesto Redrejo-Rodríguez
2018年01月05日 12430 阅读
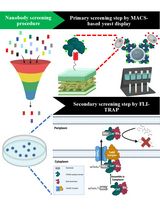
基于蛋白筛选策略从合成文库中分离抗原特异性纳米抗体:结合 MACS 的酵母展示筛选与 FLI-TRAP 方法
Apisitt Thaiprayoon [...] Dujduan Waraho-Zhmayev
2026年01月20日 402 阅读
Abstract
This protocol was developed to qualitatively and quantitatively detect protein-protein interactions in Escherichia coli by Förster Resonance Energy Transfer (FRET). The described assay allows for the previously impossible in vivo screening of periplasmic protein-protein interactions. In FRET, excitation of a donor fluorescent molecule results in the transfer of energy to an acceptor fluorescent molecule, which will then emit light if the distance between them is within the 1-10 nm range. Fluorescent proteins can be genetically encoded as fusions to proteins of interest and expressed in the cell and therefore FRET protein-protein interaction experiments can be performed in vivo. Donor and acceptor fluorescent protein fusions are constructed for bacterial proteins that are suspected to interact. These fusions are co-expressed in bacterial cells and the fluorescence emission spectra are measured by subsequently exciting the donor and the acceptor channel. A partial overlap between the emission spectrum of the donor and the excitation spectrum of the acceptor is a prerequisite for FRET. Donor excitation can cross-excite the acceptor for a known percentage even in the absence of FRET. By measuring reference spectra for the background, donor-only and acceptor-only samples, expected emission spectra can be calculated. Sensitized emission for the acceptor on top of the expected spectrum can be attributed to FRET and can be quantified by spectral unmixing.
Keywords: Bacteria (细菌)Background
Determining how and which proteins interact to sustain life is at the core of molecular biology research. Many in vitro methods exist but may result in false positives as the interactions are taken out of their biological context. An in vivo method is crucial for the verification of in vitro obtained results. Moreover, in vivo methods allow for the observation of the often, dynamic processes that take place in the cell. Fluorescent proteins (FP)s are ideal for in vivo experiments as they can be fused to proteins of interest making it possible to microscopically track the localization and dynamics of the fused proteins in living cells. Energy can be transferred between FPs by Förster Resonance Energy Transfer (FRET) provided they have compatible physical properties. The emission spectrum of a donor FP must overlap with the excitation spectrum of an acceptor FP and the distance between them should be less than 10 nm. The larger the spectral overlap and the smaller the distance between the donor and acceptor FP the more FRET can occur. The stringent distance dependence for FRET is ideal to detect direct protein-protein interactions as they also occur in the nanometer range, whereas indirect protein interactions are usually on a larger distance scale and not detectable by FRET. For this reason, it gives very few false positives but due to the physical requirements of the involved fluorophores, negative results could be false negatives. The advantage of using FPs for FRET measurements is that the chromophore is enclosed in a protein barrel and therefore much better protected from the environment than chemical chromophores. The fluorescence of the latter is extremely sensitive to environmental changes such as pH, salt, and solvents. Consequently, they often cannot be used reliably to measure FRET in a complicated environment like the living cell. A drawback of the 3-4 nm in diameter protein-barrel that encloses the chromophore in FPs is that it reduces the detectable distance between proteins to 1-6 nm. Since 5-7 nm is the typical size of proteins, it usually does not pose a problem, but can again result in false negatives. We have used a spectroscopy based FRET method to analyze the interaction between proteins in the cytoplasm and in the periplasm of Escherichia coli (Alexeeva et al., 2010; Fraipont et al., 2011; van der Ploeg et al., 2013 and 2015; Meiresonne et al., 2017) and showed that the technique is suitable for mode of action studies of antibiotics (van der Ploeg et al., 2015) and medium throughput screening (Meiresonne et al., 2017) using various FP FRET pairs. This bio-protocol describes how to perform our FRET assay in bacteria to determine protein-protein interactions by spectral unmixing. Although this protocol is written for E. coli, the method of spectral unmixing is suitable for any organism, provided fusion proteins can be expressed in the species and fluorescence spectra can be collected. The protocol describes how to perform FRET experiments by 3 approaches: 1) Measurements on fixed cell using a fluorometer, the initial determination of protein interactions. 2) Measurements on fixed cells in the plate reader for the faster detection of already confirmed protein interactions that provide clear fluorescence signals. And 3) for established interactions, measurements on living cells grown in the plate reader. For instance, to monitor the real time inhibition of a particular interaction.
Materials and Reagents
- Sterile straight neck culturing flasks (DWK Life Sciences, DURAN, catalog number: 2177124 ) with aluminum anodized metal blue cap (DWK Life Sciences, DURAN, catalog number: 2901324 )
- 50 ml sterile Falcon tubes (SARSTEDT, catalog number: 62.657.254 )
- 2 ml sterile Eppendorf tubes (Greiner Bio One International, catalog number: 623201 )
- 1.5 ml sterile Eppendorf tubes (Greiner Bio One International, catalog number: 616201 )
- Disposable pipet tips 200 µl (Ultratip, Greiner Bio One International, catalog number: 739290 )
- Disposable pipet tips 1,000 µl (Ultratip, Greiner Bio One International, catalog number: 686290 )
- Wash bottles (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 2401-0500 )
- Glass-bottomed black walled 96 wells plate (Porvair Sciences, catalog number: 324002 )
- Black walled clear bottomed 96 wells plastic plate (Greiner Bio One International, catalog number: 675096 )
- Stirred Quartz cuvettes (Hellma, catalog number: 119004F-10-40 )
- Lens cleaning tissue (Fischer Scientific, FisherbrandTM, catalog number: 10605955 )
Note: This product has been discontinued. - Filtropur S 0.2 µm filter (SARSTEDT, catalog number: 83.1826.001 )
- Ethanol 70% in lab wash bottle (VWR, Technisolv®, catalog number: 83801.360 )
- Ethanol 96% in lab wash bottle (VWR, Technisolv®, catalog number: 83804.360 )
- H2O in lab wash bottle
- 1 M β-D-1-thiogalactopyranoside (IPTG) (Duchefa Biochemie, catalog number: I1401 )
- Hellmanex III (Hellma, catalog number: 9-307-011-4-507 )
- Bacto tryptone (Duchefa Biochemie, catalog number: T1332 )
- Yeast extract (Duchefa Biochemie, catalog number: Y1333 )
- Sodium chloride (NaCl) (Merck, catalog number: 106404 )
- Glucose (Duchefa Biochemie, catalog number: G0802 )
- Thiamine hydrochloride (Sigma-Aldrich, catalog number: T4625-25G )
- L-Lysine monohydrochloride (Sigma-Aldrich, catalog number: L8662 )
- Ampicillin stock solution 100 mg ml-1 (Sigma-Aldrich, catalog number: A9518 ) in 50% ethanol
- Chloramphenicol stock solution 25 mg ml-1 (Duchefa Biochemie, catalog number: C0113 ) in 50% ethanol
- di-Potassium hydrogen phosphate (K2HPO4·3H2O) (VWR, catalog number: 26932.290 )
- Potassium dihydrogen phosphate (KH2PO4) (Merck, catalog number: 104873 )
- Ammonium sulfate, (NH4)2SO4 (Sigma-Aldrich, catalog number: A6387 )
Note: This product has been discontinued. - Magnesium sulfate heptahydrate (MgSO4·7H2O) (Carl Roth, catalog number: T888.1 )
- Iron(II) sulfate heptahydrate (FeSO4·7H2O) (Sigma-Aldrich, catalog number: 215422 )
- Calcium nitrate tetrahydrate, Ca(NO3)2·4H2O (Sigma-Aldrich, catalog number: C1396 )
- L-Arginine (Sigma-Aldrich, catalog number: A8094 )
- L-Glutamine (Sigma-Aldrich, catalog number: G8540 )
- Thymidine (Sigma-Aldrich, catalog number: T1895 )
- Uracil (Sigma-Aldrich, catalog number: U1128 )
- Potassium chloride (KCl) (VWR, catalog number: 71003-522 )
- di-Sodium hydrogen phosphate dihydrate (Na2HPO4·2H2O) (Merck, catalog number: 106580 )
- Glutaraldehyde 25% (Merck, catalog number: 1042390250 )
- Formaldehyde ≥ 36.0% (Sigma-Aldrich, catalog number: 47608 )
- Lysine (Sigma-Aldrich, catalog number: L8662 )
- TY medium (see Recipes)
- 20% glucose solution (see Recipes)
- GB1 medium (see Recipes)
- MM1-10x
- MM2-10x
- MM3-100x
- MM4-100x
- Vitamin B1 stock solution (4 mg ml-1)
- Lysine stock solution (20 mg ml-1)
- MM1-10x
- Phosphate buffered saline (PBS 1x) (see Recipes)
- Formaldehyde and glutaraldehyde (FAGA) (see Recipes)
Equipment
- P20, P200 & P1000 Pipetman pipettes (Gilson, catalog number: F167300 )
- Gyrotory water bath shaker Model G76 (Eppendorf, New Brunswick Scientific, model: Model G76 or GFL-Gesellschaft für Labortechnik, catalog number: 1092 ) with holders for culturing flasks
- Photospectrometer (Biochrom, model: Libra S22 )
- Compressed air source
- Water purification system (Purelab flex with ELGA LC197-Biofilter from ELGA LabWater, Laner End, HP14 3BY, UK)
Note: All H2O in the protocol indicates water of 11.5 MΩ from Water purification system. - Centrifuge for 50 ml Falcon tubes (Eppendorf, model: 5804 R )
- Centrifuge for 1.5 and 2.0 ml Eppendorf tubes (Eppendorf, model: 5424 R )
- Spectrofluorometer Quantamaster 2000-4 (Photon Technology International, NJ) with a red-optimized set-up: R928P PMT tube (185-900 nm), 500 nm blaze (1,200 line/mm) both excitation and emission gratings
Note: A PMT that will reliably detect fluorescence spectra up until 700 nm will be suitable for the described protocol. - Optical filters:
- 500/10 nm BrightLine® single-band bandpass filter (Semrock, catalog number: FF01-500/10 )
- ET510LP (Chroma Technology, catalog number: ET510lp )
- 514/3 nm BrightLine® single-band bandpass filter (Semrock, catalog number: FF01-514/3 )
- 515 nm blocking edge BrightLine® long-pass filter (Semrock, catalog number: FF01-515/LP )
- HQ541/12 (Chroma Technology, catalog number: HQ541/12 )
Note: This product has been discontinued. Filters with similar transmission properties can be used. - E550LP (Chroma Technology, catalog number: E550lp )
- 587/11 nm BrightLine® single-band bandpass filter (Semrock, catalog number: FF01-587/11 )
- HQ600LP (Chroma Technology, catalog number: HQ600LP )
Note: This product has been discontinued.
- 500/10 nm BrightLine® single-band bandpass filter (Semrock, catalog number: FF01-500/10 )
- Fluorescence scanning plate reader (BioTek Instruments, model: SynergyTM Mx , catalog number: SMATBC)
- Autoclave (SANYO, catalog number: MLS-3780 )
Software
- Fluorescence spectrophotometer software (PTI acquisition software FeliX32 version 1.2 Build 56)
- Microplate reader software Gen5 version 2.00 (Biotek)
- Excel (Microsoft)
Procedure
文章信息
版权信息
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Meiresonne, N. Y., Alexeeva, S., van der Ploeg, R. and den Blaauwen, T. (2018). Detection of Protein Interactions in the Cytoplasm and Periplasm of Escherichia coli by Förster Resonance Energy Transfer. Bio-protocol 8(2): e2697. DOI: 10.21769/BioProtoc.2697.
分类
微生物学 > 体内实验模型 > 细菌
生物化学 > 蛋白质 > 相互作用 > 蛋白质-蛋白质相互作用
分子生物学 > 蛋白质 > 蛋白质-蛋白质相互作用
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link