- EN - English
- CN - 中文
Immunogold Electron Microscopy of the Autophagosome Marker LC3
自噬体标志物LC3的免疫金电镜观察
发布: 2017年12月20日第7卷第24期 DOI: 10.21769/BioProtoc.2648 浏览次数: 13285
评审: Vikash VermaAswad KhadilkarMartin SachseAnonymous reviewer(s)
Abstract
Even though autophagy was firstly observed by transmission electron microscopy already in the 1950s (reviewed in Eskelinen et al., 2011), nowadays this technique remains one of the most powerful systems to monitor autophagic processes. The autophagosome, an LC3-positive double membrane structures enclosing cellular materials, represents the key organelle in autophagy and its simple visualization and/or numeration allow to draw important conclusions about the autophagic flux. Therefore, the accurate identification of autophagosomes is crucial for a comprehensive and detailed dissection of autophagy. Here we present a simple protocol to identify autophagosomes by transmission electron microscopy coupled to immunogold labeling of LC3 starting from a relatively low cell number, which we recently developed to follow the autophagic pathway during viral-mediated human carcinogenesis.
Keywords: Autophagy (自噬)Background
The autophagosome represents the key structure in macroautophagy, a catabolic degradation system for cellular cytosolic constituents. Macroautophagy (or simply autophagy) is initiated by the formation of the phagophore, a membrane able to expand itself engulfing organelles and proteins that finally closes around sequestered components forming an organelle known as autophagosome. Next, during the maturation process, the autophagosome can fuse with lysosomes to form autolysosomes where trapped materials are degraded and recycled back by pumps located in the lysosomal limiting membrane (Glick et al., 2010).
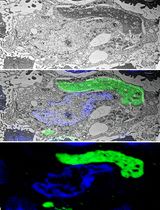
Since malfunction of autophagy has been linked to a variety of human disease, viral infection, neurodegeneration, immune function, and cancer (Schneider and Cuervo, 2014), a more comprehensive and detailed dissection of autophagy is increasingly required. Indeed, numerous methodologies to study autophagy have been developed so far (Klionsky et al., 2016), and most of them need to be simultaneously performed in order to correctly define how autophagy is working in a particular experimental system. Among them, transmission electron microscopy (TEM) coupled with specific immunogold labeling (immunogold electron microscopy-IEM) remains one of the most accurate methods for autophagy detection because of its exquisite ability to provide fine details of specific ultracellular structures. Indeed, in addition to their peculiar morphology (Yla-Anttila et al., 2009), autophagosomes are also characterized by the presence of microtubule-associated protein 1 light chain 3, or LC3, which is generally recruited on autophagic membranes after lipidation with phosphatidylethanolamine (LC3-II) during autophagic activation (Kabeya et al., 2000). Therefore, TEM coupled with labeling of cells with anti-LC3 antibody and gold probes conjugated to a secondary antibody allows the unequivocal identification of the autophagosome, and indirectly, permits to monitor autophagic flux.
Several approaches are commonly used for the immunogold labelling of cultured cells: pre-embedding (immunolabeling before resin embedding) and post-embedding methods (immunolabeling after resin embedding) including also Tokuyasu cryosections, where cryoprotected embedded specimens are firstly frozen by immersion in liquid nitrogen and then cut with cryo-ultramicrotome at -120 °C before immunolabelling (Tokuyasu, 1980; Eskelinen et al., 2002; Jager et al., 2004). However, both techniques require at least 3 x 106 cells to obtain an adequate pellet for sample preparation, inferring that initial cell number is a critical factor to consider during planning of IEM experiments.
Therefore, we describe herein a pre-embedding protocol for autophagosome detection starting from small cell numbers (less than 0.1 x 106 cells), which allows performing IEM analysis in primary cells characterized by low replicative potential. We recently used this protocol to detect LC3-positive autophagosomes on populations of primary human keratinocytes transduced with retroviral vectors relevant to viral carcinogenesis (Mattoscio et al., 2017). In principle, this methodology could be applied to detect other antigens of interest, since we used the same protocol also to study the subcellular localization of the SUMO-conjugating enzyme UBC9 (Mattoscio et al., 2017), and may be used with different cellular species for which the initial cell number is also a limiting factor.
Materials and Reagents
- 13 mm Nunc Thermanox coverslips (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 150067 )
- 24-well tissue culture-treated plates (Corning, Costar®, catalog number: 3524 )
- Eppendorf® Safe-Lock microcentrifuge tubes (Sigma-Aldrich, catalog number: T9661)
Manufacturer: Eppendorf, catalog number: 022363204 . - Edge razor blade (Sigma-Aldrich, catalog number: Z740504 )
- 200 mesh grids (Sigma-Aldrich, catalog number: G5151 )
- Chang Monolayer Molds (Electron Microscopy Sciences, catalog number: 70920 )
- Ethanol BioUltra, for molecular biology (Sigma-Aldrich, catalog number: 51976 )
- Dulbecco’s phosphate-buffered saline, calcium, magnesium (Sigma-Aldrich, catalog number: D8662 )
- Paraformaldehyde (Sigma-Aldrich, catalog number: P6148 )
Note: To prepare 4% paraformaldehyde (PFA) solution, in a fume hood dissolve 4 g of PFA in 90 ml of water at 60 °C and few drops of 1 N NaOH (until the solution appears without PFA precipitate), then remove from heating, add 10 ml 10x PBS, fill up with water up to 100 ml, and cool. Do not heat solution above 70 °C since PFA will break down at higher temperatures. Use the solution immediately or store for 1 week at 4 °C or at -20 °C for a longer time. - Glutaraldehyde (Sigma-Aldrich, catalog number: G7776 )
- Quenching solution: 50 mM glycine (Sigma-Aldrich, catalog number: 410225 ) in PBS
- Primary antibody: rabbit polyclonal anti-LC3 (Novus Biologicals, catalog number: NB100-2331 )
- Washing buffer (0.1% BSA, 0.1% saponin in PBS)
- Secondary antibody conjugated with gold: 1.4 nm Nanogold IgG goat anti rabbit IgG (Nanoprobes, catalog number: 2003 )
- Gold enhancement solution (GoldEnhanceTM EM) (Nanoprobes, catalog number: 2113 ), made by four different components: A (enhancer), B (activator), C (initiator) and D (buffer)
- Uranyl acetate (Electron Microscopy Sciences, catalog number: 22400 )
Note: Uranyl acetate is mildly reactive and highly toxic by ingestion. Please dispose of waste according to your institutional policy. - Embed-812 resin kit (Electron Microscopy Sciences, catalog number: 14120 )
Notes:- This product has been discontinued and replaced by EMbed 812.
- For a better preservation, mix Embed-812 (32 g) with Dodecenyl succinie anhydride (DDSA) (16 g) and NMA (Nadic Methyl Anhydride) (17.4 g). Mix well, add 0.8 g of the accelerator DMP30 (2,4,6-tri (dimethylaminomethyl) phenol), mix and store in 20 ml syringe at -20 °C. Warm to room temperature before use.
- This product has been discontinued and replaced by EMbed 812.
- Hydrofluoric acid (Fisher Scientific, catalog number: 14-650-236)
Manufacturer: Avantor Performance Materials, J.T. Baker, catalog number: 9560-01 . - Blocking buffer (see Recipes)
- Bovine serum albumin (BSA, IgG-Free and Protease-Free) (Jackson ImmunoResearch, catalog number: 001-000-162 )
- Normal goat serum (Jackson ImmunoResearch, catalog number: 005-000-121 )
- Ammonium chloride (Sigma-Aldrich, catalog number: A9434 )
- Saponin (Sigma-Aldrich, catalog number: 47036 )
- Phosphate buffer (Sigma-Aldrich, catalog number: P3619 )
- Sodium chloride (Sigma-Aldrich, catalog number: S7653 )
- Bovine serum albumin (BSA, IgG-Free and Protease-Free) (Jackson ImmunoResearch, catalog number: 001-000-162 )
- Reduced osmium tetroxide (see Recipes)
- Sato lead solution (see Recipes)
- Lead citrate tribasic (Sigma-Aldrich, catalog number: 15326 )
- Lead nitrate (Merck, catalog number: 1073980100 )
- Sodium citrate tribasic dehydrate (Sigma-Aldrich, catalog number: S4641 )
- Lead citrate tribasic (Sigma-Aldrich, catalog number: 15326 )
Equipment
- Fume hood
- 60 °C Oven (Memmert, model: UN30 )
- Ultramicrotome (Leica Microsystem, model: Leica EM UC7 )
- Transmission Electron microscope (Carl Zeiss, model: Leo 912AB ) equipped with a slow-scan Proscan camera (ProScan)
- Rocking shaker
Procedure
文章信息
版权信息
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Mattoscio, D., Raimondi, A., Tacchetti, C. and Chiocca, S. (2017). Immunogold Electron Microscopy of the Autophagosome Marker LC3. Bio-protocol 7(24): e2648. DOI: 10.21769/BioProtoc.2648.
分类
细胞生物学 > 细胞染色 > 蛋白质
免疫学 > 宿主防御 > 综合
细胞生物学 > 细胞成像 > 电子显微镜
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link