- EN - English
- CN - 中文
Protocol for Construction of a Tunable CRISPR Interference (tCRISPRi) Strain for Escherichia coli
大肠埃希杆菌可调CRISPR干扰(tCRISPRi)菌株的构建实验方案
发布: 2017年10月05日第7卷第19期 DOI: 10.21769/BioProtoc.2574 浏览次数: 10837
评审: Anonymous reviewer(s)

相关实验方案
氨基甲酰转移酶测定: 5-羟甲基胞嘧啶 (5hmC) 到 5-氨基甲酰氧基甲基胞嘧啶 (5cmC) 的体外修饰
Weiwei Yang [...] Laurence Ettwiller
2022年09月05日 1982 阅读
Abstract
We present a protocol for construction of tunable CRISPR interference (tCRISPRi) strains for Escherichia coli. The tCRISPRi system alleviates most of the known problems of plasmid-based expression methods, and can be immediately used to construct libraries of sgRNAs that can complement the Keio collection by targeting both essential and nonessential genes. Most importantly from a practical perspective, construction of tCRISPRi to target a new gene requires only one-step oligo recombineering. Additional advantages of tCRISPRi over other existing CRISPRi methods include: (1) tCRISPRi shows significantly less than 10% leaky repression; (2) tCRISPRi uses a tunable arabinose operon promoter and modifications in transporter genes to allow a wide dynamic range with graded control by arabinose inducer; (3) tCRISPRi is plasmid free and the entire system is integrated into the chromosome; (4) tCRISPRi strains show desirable physiological properties.
Keywords: CRISPR interference (CRISPR干扰)Background
Various CRISPR interference systems have been developed for use in organisms from bacteria to eukaryotes. For those who are considering to use CRISPRi for bacteria, we provide the following background information on our tCRISPRi system (Li et al., 2016) and its comparison with other CRISPRi systems.
Morgan-Kiss et al. (2002) developed the plasmid-based, dose-inducible promoter pBAD. Their system allows tunable expression of a protein from the pBAD promoter, dependent upon arabinose levels. The arabinose transporter genes araE and araFGH are inactive in the strain. Their strain also has two copies of lacY; the wild-type lacY on the chromosome and a mutant lactose transporter lacY A177C on a plasmid. The LacY A177C function allows arabinose to diffuse into the cell, and thus, the pBAD induction level is precisely controlled by the concentration of the supplied arabinose in the medium (Morgan-Kiss, 2002).
Our tCRISPRi strain contains only the mutant gene lacY A177C (Morgan-Kiss, 2002), which is expressed from the lac operon constitutively because the lacI repressor gene is deleted. Our strain also has gene deletions of araE and araFGH. LacY A177C is the only arabinose transporter in the cell allowing for better control of the PBAD promoter and tunable repression by tCRISPRi.
A recent study by Peters et al. (2016) showed the power of CRISPR-based knockdown methods for studying essential genes in Bacillus subtilis. Their sgRNA libraries were cloned via inverse PCR, and dCas9 was under a xylose-inducible promoter. In contrast, our tCRISPRi system for E. coli uses one-step recombineering to make a tCRISPRi strain. The PBAD promoter in the present work shows about 7.5% leaky expression, whereas the B. subtilis CRISPRi shows approximately 33% leakiness. Another important pioneering CRISPRi system was designed by the Marraffini group (2013), who used a plasmid-based system. We compare our tCRISPRi with these other two systems in Table 1. To see an example of applications of tCRISPRi to essential cell cycle genes, see Si et al. (2017).
Table 1. Comparison of different CRISPR interference system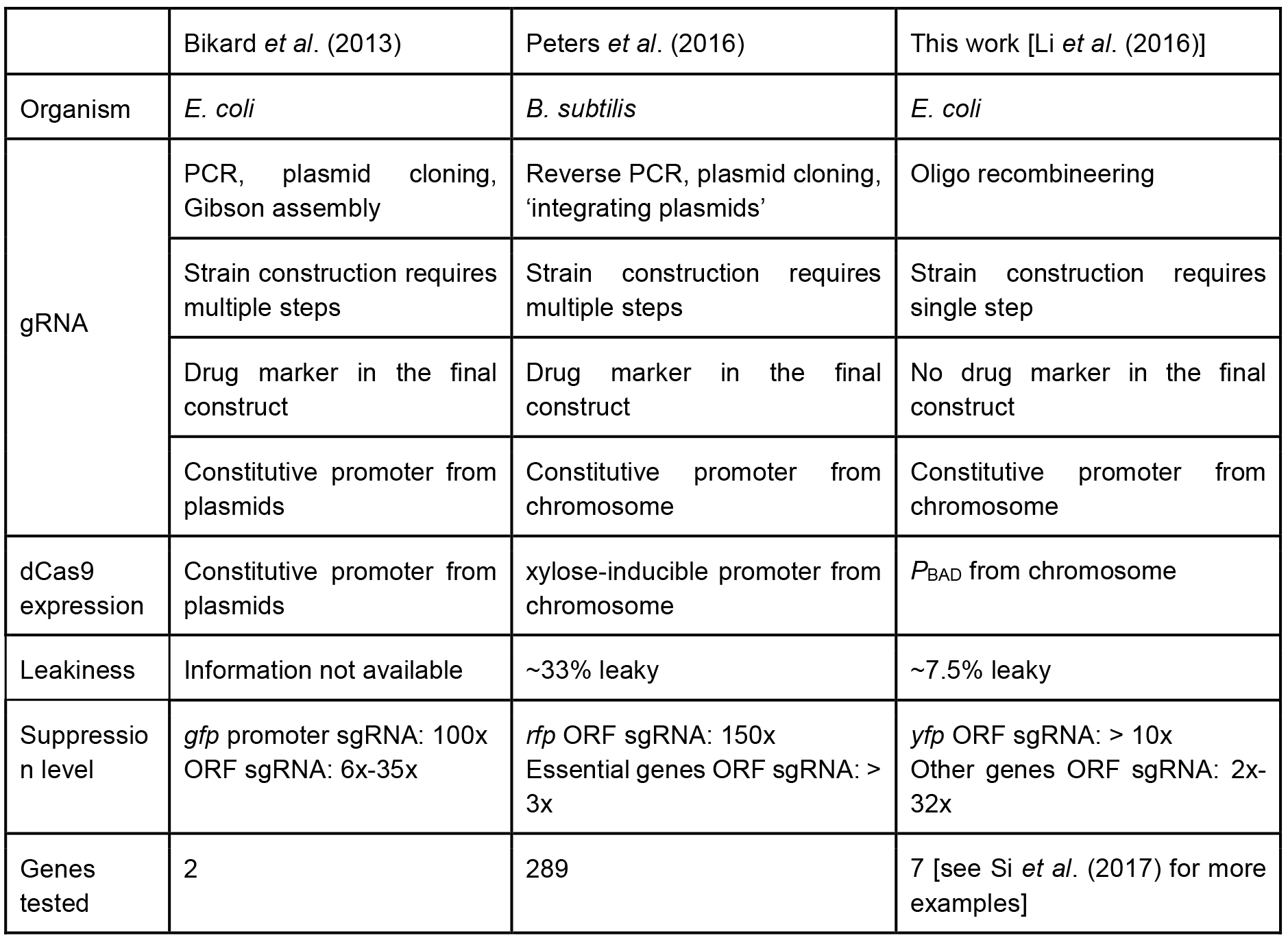
Materials and Reagents
- 1.5 ml microcentrifuge tubes
- Pipette tips (10 μl , 200 μl, 1,000 μl) (Genesee Scientific, catalog numbers: 24-121RL )
- Pipette tips (10 μl , 200 μl, 1,000 μl) (Genesee Scientific, catalog numbers: 24-150RL )
- Pipette tips (10 μl , 200 μl, 1,000 μl) (Genesee Scientific, catalog numbers: 24-165RL )
- 15 ml culture tubes
- Electroporation cuvettes, 1 mm gap (Bio-Rad Laboratories, catalog number: 1652089 )
- Petri dishes
- SJ_XTL219 strain available from Addgene (Addgene, catalog number: 86400 )
- Ultrapure water
- Antibiotics:
Hygromycin (50 mg/ml) (Thermo Fisher Scientific, InvitrogenTM, catalog number: 10687010 )
Tetracycline (12.5 mg/ml) (Sigma-Aldrich, catalog number: T3383 ) - PCR purification kit (QIAGEN, catalog number: 28104 )
- Phusion high fidelity polymerase (New England Biolabs, catalog number: M0530L )
- L(+)Arabinose (EMD Millipore, Calbiochem, catalog number: 178680 )
- Gel extraction kit (QIAGEN, catalog number: 28704 )
- Tryptone (EMD Millipore, catalog number: 1.07213.1000 )
- Yeast extract (Sigma-Aldrich, catalog number: Y1625-1KG )
- Sodium chloride (NaCl) (Fisher Bioreagents, catalog number: BP3581 )
- LB agar (Fisher Bioreagents, catalog number: BP1425 )
- Luria broth (LB) (see Recipes)
- LB agar plate (see Recipes)
- Sucrose plates (see Recipes)
Equipment
- Pipettes (that can accommodate pipette tips in 2 above)
- Milli-Q water purification system (Millipore, catalog number: Z00Q0V0WW )
- Autoclave (AMSCO, model: 3041-S )
- Incubator and shaker (32 °C and 42 °C) (Eppendorf, New BrunswickTM, model: Innova® 3100 , catalog number: M1231-0000)
- Microcentrifuge (Eppendorf, catalog number: 5424 )
- Electroporator (Bio-Rad Laboratories, catalog number: 1652100 )
- Gel electrophoresis chamber (Bio-Rad Laboratories, catalog number: 1704406 )
- Thermal cycler (Bio-Rad Laboratories, model: C1000 TouchTM, catalog number: 1851148 )
- Nikon Inverted Microscope Eclipse Ti-E (Nikon, model: Eclipse Ti-E , catalog number: MEA53100) equipped with an Andor Neo sCMOS camera (Nikon, model: Neo 5.5 , catalog number: 77026046)
Software
- Nikon NIS-Elements Advanced Research software
Procedure
文章信息
版权信息
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Li, X., Sou, C. and Jun, S. (2017). Protocol for Construction of a Tunable CRISPR Interference (tCRISPRi) Strain for Escherichia coli. Bio-protocol 7(19): e2574. DOI: 10.21769/BioProtoc.2574.
分类
分子生物学 > DNA > DNA 修饰
微生物学 > 微生物遗传学 > 基因图谱和克隆
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link