- EN - English
- CN - 中文
Detection of Pathogens and Ampicillin-resistance Genes Using Multiplex Padlock Probes
使用多重锁式探针检测病原体和氨苄西林耐药基因
发布: 2017年08月20日第7卷第16期 DOI: 10.21769/BioProtoc.2504 浏览次数: 10257
评审: Modesto Redrejo-RodriguezAnonymous reviewer(s)
Abstract
Diagnostic assays for pathogen identification and characterization are limited either by the number of simultaneously detectable targets, which rely on multiplexing methods, or by time constraints due to cultivation-based techniques. We recently presented a 100-plex method for human pathogen characterization to identify 75 bacterial and fungal species as well as 33 clinically relevant β-lactamases (Barišić et al., 2016). By using 16S rRNA gene sequences as barcode elements in the padlock probes, and two different fluorescence channels for species and antibiotic resistance identification, we managed to cut the number of microarray probes needed by half. Consequently, we present here the protocol of an assay with a runtime of approx. 8 h and a detection limit of 105 cfu ml-1. A total of 89% of β-lactamases and 93.7% of species were identified correctly.
Keywords: Multiplex detection (多重检测)Background
β-Lactamases are a class of antibiotic resistance genes which provide resistance to β-lactam antibiotics, which structurally mimic D-alanyl-D-alanine, a component of the bacterial cell wall and thereby inhibit bacterial cell wall synthesis. β-Lactamases are able to hydrolyze the central component of β-lactam antibiotics, the β-lactam ring, and render them useless (Kong et al., 2010). Today, over 1,000 β-lactamases are described and a huge potential environmental reservoir exists (Bush, 2010; Brandt et al., 2017). β-Lactamases are ancient enzymes and we classify them as class A, C, and D (serine β-lactamases) with a serine catalytic site, or as class B (metallo-β-lactamases) whose active center is zinc-dependent (Hall and Barlow, 2003 and 2004). Despite their high phylogenetic age, the serine β-lactamases probably share a common ancestor and acquired a high number of SNPs due to a permanent selection pressure. Additionally, to β-lactamases, more than 500 other antibiotic resistance genes exist (Zankari et al., 2012).
Current multiplexing methods reduce the number of simultaneously detectable targets while cultivation-dependent techniques are limited by time constraints. Given these facts and the high number of pathogens of clinical importance, new methods are needed for the fast characterization and identification of pathogens, virulence factors and antibiotic resistance genes.
The gold standard of infection diagnostics takes 2-3 days and is cultivation-dependent (Marik, 2014). Additionally, PCR methods provide results at a high sensitivity and low cost, but remain impractical due to the high number of clinically relevant targets (Mussap et al., 2007; Wellinghausen et al., 2009). The current multiplex-PCR protocols are not suitable for a high number of targets and the limitations can only be overcome by microfluidic-based assays, which run a high number of analyses in parallel.
Padlock probes are linear DNA probes, which upon annealing, circularize and are then used for rolling circle amplifications (Nilsson et al., 1994; Hardenbol et al., 2005). They allow for a higher number of multiplexing and can easily be integrated into established PCR-based assays. Recently, we presented a 100-plex method based on padlock probes for pathogen characterization (75 bacterial and fungal species) as well as 33 clinically important β-lactamases (Barišić et al., 2016). By adapting this method from our previous work (Barišić et al., 2013), we increased the sensitivity and specificity of the assay and we managed to cut down the number of microarray probes needed by half by using 16S rRNA sequences as barcode elements in the padlock probes and two different fluorescence channels for species identification and antibiotic resistance characterization. Here, we present an assay to overcome time limitations and to increase the number of detectable targets. Our assay allows for the detection of up to 105 cfu ml-1 in a total of 8 h. We were able to retrieve and correctly characterize 89% of β-lactamases and to identify 93.7% of all species.
Materials and Reagents
- Filter tips 10 µl, 20 µl, 200 µl and 1,250 µl (e.g., Biozym, catalog numbers: VT0200 , VT0220 , VT0240 and VT0270 )
- Safe-Lock tubes 1.5 ml (Eppendorf, catalog number: 022363204 )
- Falcon 15 ml conical centrifuge tubes (Corning, catalog number: 352196 )
- 2 ml screw-cap tube (e.g., Roche Molecular Systems, catalog number: 03358941001 )
- Ritter Riplate 384 well plate PP (Ritter, catalog number: 43001-0035 )
- Glass beads, acid-washed, 150-212 μm (Sigma-Aldrich, catalog number: G1145 )
- Glass beads, acid-washed, 425-600 μm (Sigma-Aldrich, catalog number: G8772 )
- Vantage Silylated Aldehyde Slides (CEL Associates, catalog number: VSS-25 )
- LifterSlip mSeries cover slips for microarray slides, 55 µl (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 25X60IM5439001LS )
- PCR tubes 0.2 ml (Eppendorf, catalog number: 0030124537 )
- Millex-GV 0.22 µm syringe filter units (Merck, catalog number: SLGV033RS )
- 50 ml tubes (e.g., Corning, catalog number: 430829 )
- Disposable syringes, e.g., Omnifix 50 ml LL (B. Braun Medical, catalog number: 8508577FN )
- The multiplex PCR primers (66 in total, Table S1) targeting the β-lactamase genes were ordered from Microsynth (Balgach, Switzerland) (Note 3)
- The padlock probes (66 in total, Figure 1, Table S2) targeting the β-lactamases were ordered from Integrated DNA Technologies (Coralville, IA, USA)
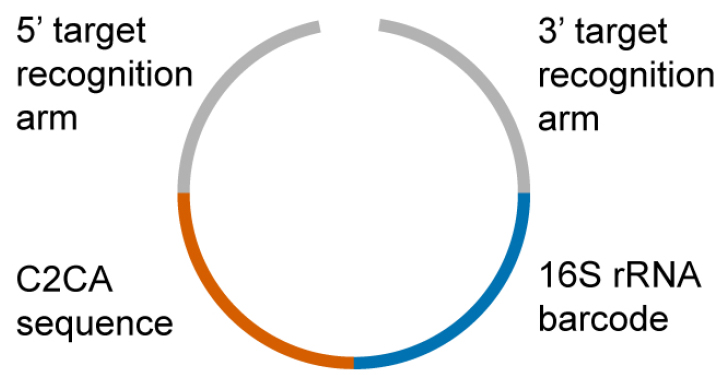
Figure 1. Schematic illustration of a padlock probe. The 3’ and the 5’ target recognition arms bind to the multiplex PCR products of the β-lactamase genes. During the binding process, the padlock probes are circularized and subsequently ligated. The maximum distance of the padlock probe binding region to the 3’ or 5’-ends of the PCR product should not exceed 200 base pairs. Since the padlock probes gets concatenated with the PCR product upon the ligation reaction, longer distances cause an inhibition of subsequent RCAs. The C2CA sequence is needed for the circle-to-circle amplification and comprises an AluI restriction site for monomerization of the amplification products. The barcode sequence is derived from the 16S rRNA gene. This allowed us to halve the number of microarray probes because the C2CA products and 16S rRNA gene PCR products are detected on the same microarray probe but in different fluorescence channels. - The 5’-amino-modified microarray probes (274 in total, Table S3) were ordered from Microsynth (Balgach, Switzerland) (Note 3)
- Nuclease-free water for PCR application comes with the Mastermix 16S Basic kit or can be purchased separately (e.g., Fresenius Kabi, Aqua bidest. ‘Fresenius’, no catalog number)
- Ultrapure water, hereafter simply referred to as water or H2O (Note 1)
- The universal bacterial 16S rRNA primers 45f++ (5’-GCYTAAYACATGCAAGTCGARCG-3’) and 783R (5’-TGGACTACCAGGGTATCTAATCCT-3’) were ordered from Integrated DNA Technologies (Coralville, IA, USA)
- The fungal 18S rRNA (ITS region) primers ITS3 (5’-GCATCGATGAAGAACGCAGC-3’) and ITS4+ (5’-TCCT-CCGCTTATTGATATGCTTAAGT-3’) were ordered from Integrated DNA Technologies (Coralville, IA, USA)
- The circle-to-circle amplification (C2CA) oligonucleotides C2CA- (5’-TACTCGAGGAGCTGCATACAC-3’) and C2CA+ (5’-GTGTATGCAGCTCCTCGAGTA-3’) were ordered from Integrated DNA Technologies (Coralville, IA, USA)
- The Cy5-labelled hybridization control (complimentary to ‘Bsrev’) (5’-Cy5-AAGCTCACTGGCCGTCGTTTTAAA-3’) was ordered from Microsynth (Balgach, Switzerland)
- T4 DNA ligase (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: EL0011 )
- T4 polynucleotide kinase (10 U/µl) supplied with 10x reaction buffer A (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: EK0031 )
- ATP solution (100 mM) (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: R0441 )
- Mastermix 16S Basic, DNA-free (Molzym, catalog number: S-040-0250 ) containing a 2.5x complete master mix, Moltaq 16S DNA polymerase and PCR-grade water (Note 2)
- dNTP mix (10 mM each) (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: R0192 )
- Cy5-dCTP (1 mM solution) (GE Healthcare, catalog number: PA55021 )
- VentR (exo-) DNA polymerase (New England Biolabs, catalog number: M0257S ) supplied with 10x ThermoPol reaction buffer and 100 mM MgSO4
- ExpressHyb hybridization solution (Takara Bio, Clontech, catalog number: 636831 )
- Ampligase thermostable DNA ligase (5 U/µl) and Ampligase 10x reaction buffer (Epicentre, catalog number: A32750 )
- Bovine serum albumin (BSA), molecular biology grade (New England Biolabs, catalog number: B9000S )
- phi29 DNA polymerase supplied with 10x phi29 DNA polymerase reaction buffer (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: EP0091 )
- AluI restriction enzyme (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: ER0011 )
- Atto532-dCTP (MoBiTec, Göttingen, Germany)
- SDS solution 10% for molecular biology (AppliChem, catalog number: A0676 )
- Tryptic soy broth, also referred to as CASO medium (Casein-peptone soymeal-peptone broth) (Merck, catalog number: 105459 )
- PBS (10x), pH 7.2 (Thermo Fisher Scientific, GibcoTM, catalog number: 70013 )
- Betaine monohydrate (Sigma-Aldrich, catalog number: B2754 )
- UltraPure SSC, 20x (Thermo Fisher Scientific, InvitrogenTM, catalog number: 15557036 )
- CASO medium (see Recipes)
- 1x phosphate-buffered saline (PBS) (see Recipes)
- 2x spotting buffer (6x SSC, 3 M betaine) (see Recipes)
Equipment
- Pipettes (e.g., Sartorius, catalog numbers: 728020 , 728050 , 728060 and 728070 )
- Microbiological incubator shaker (e.g., IKA, model: KS 4000 i control )
- Tabletop centrifuge for 1.5 ml tubes (e.g., Eppendorf, model: 5424 )
- Roche MagNA Lyser Instrument (Basel, Switzerland)
- Thermomixer comfort (Eppendorf, Hamburg, Germany)
- Epoch Microplate spectrophotometer (Biotek, Winooski, VT, USA)
- Biosan DNA/RNA UV-cleaner box (Warren, MI, USA) (recommended, see Note 4)
- Thermal cycler (e.g., Thermo Fisher Scientific, Applied Biosystems, model: GeneAmpTM PCR System 2700 ) (Paisley, UK)
- Heraeus Megafuge 40R with a TX-750 rotor (Thermo Fisher Scientific, Thermo ScientificTM, model: HeraeusTM MegafugeTM 40R , catalog number: 75004518; TX-750 rotor: Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 75003180 ) and inserts for plates (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 75003617 ) and Falcon tubes (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 75003608 )
- Slide humidity incubation box (e.g., LabScientific, catalog number: HIC-3 )
- GeneMachines Omnigrid 100 contact arrayer (Madison, WI, USA)
- International Microarray Pin Stealth 3 SMP3 (Telechem, catalog number: SMP3 )
- Agilent SureScan DNA Microarray Scanner (Santa Clara, CA, USA)
- Autoclave
- Sartorius arium pro UV ultrapure water system (Sartorius, model: arium® pro )
Software
- GenePix Pro 6.0 Software (Molecular Devices LLC, Sunnyvale, CA, USA)
- Microsoft Excel or any other data analysis software
- ARB software package for microarray probe design (Note 3)
- Primer3 for primer design (Note 3)
Procedure
文章信息
版权信息
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Conzemius, R. and Barišić, I. (2017). Detection of Pathogens and Ampicillin-resistance Genes Using Multiplex Padlock Probes. Bio-protocol 7(16): e2504. DOI: 10.21769/BioProtoc.2504.
分类
微生物学 > 微生物遗传学 > DNA > PCR
微生物学 > 微生物-宿主相互作用 > 细菌
分子生物学 > DNA > 基因分型
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link