- EN - English
- CN - 中文
Isolation of Nuclei in Tagged Cell Types (INTACT), RNA Extraction and Ribosomal RNA Degradation to Prepare Material for RNA-Seq
从标记的细胞类型中分离细胞核、提取RNA并去除核糖体RNA以用于RNA-Seq 分析
(*contributed equally to this work) 发布: 2018年04月05日第8卷第7期 DOI: 10.21769/BioProtoc.2458 浏览次数: 16091
评审: Renate WeizbauerJuliane K IshidaHiroyuki Hirai
Abstract
Gene expression is dynamically regulated on many levels, including chromatin accessibility and transcription. In order to study these nuclear regulatory events, we describe our method to purify nuclei with Isolation of Nuclei in TAgged Cell Types (INTACT). As nuclear RNA is low in polyadenylated transcripts and conventional pulldown methods would not capture non-polyadenylated pre-mRNA, we also present our method to remove ribosomal RNA from the total nuclear RNA in preparation for nuclear RNA-Seq.
Keywords: Gene expression (基因表达)Background
Isolating specific cell types for gene expression experiments reduces the noise and increases the precision of the experiment and the number of differently expressed genes found. Various methods for cell type-specific studies are widely used, each one with their strengths and weaknesses (reviewed in Bailey-Serres, 2013). Isolating specific regulatory compartments, such as nuclei (from organelles, ribosomes, cytosol, etc.) provides further precision in dissecting the molecular events in regulation of gene expression. Here we describe a method that allows isolating nuclei of specific cell types from frozen tissue, suited for experiments where nuclear gene expression is to be studied (e.g., RNA-Seq of nuclear RNA, ATAC-Seq, ChIP-Seq, etc.). Furthermore, we describe the processing of RNA that leads to material suited to be the input for an RNA-Seq experiment. The protocols described here were used with rice root tissue (Oryza sativa cv. Nipponbare) (Reynoso et al., 2018), but they are based on previous protocols developed for Arabidopsis (Deal and Henikoff, 2010 and 2011) and tomato (Ron et al., 2014).
The first part of this protocol, the INTACT method (for Isolation of Nuclei Tagged in specific Cell Types) allows in vivo affinity labeling and subsequent purification of nuclei from a cell type of interest. This is achieved through cell type-specific expression of a tripartite nuclear tagging fusion protein (NTF) consisting of a nuclear envelope targeting domain, GFP, and the biotin ligase recognition peptide (BLRP). Co-expression of NTF along with the E. coli biotin ligase gene codon optimized for rice, BirA, in the cell type of interest results in the production of fluorescently labeled, biotinylated nuclei specifically in that cell type. These labeled nuclei can then be affinity purified from a crude tissue homogenate using streptavidin-coated magnetic beads, thus allowing access to RNA and chromatin from the cell type of interest.
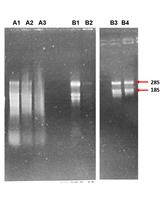
The second part of the method describes the processing of nuclear RNA to produce a sample that is suitable to be the input for RNA-Seq library preparation. Typically for eukaryotic samples, ribosomal RNA (rRNA) and organellar RNA is removed from RNA samples by polyA-pulldown methods. However, nuclear mRNA contains pre-mRNA in many stages of processing, most not polyadenylated, while many of the polyadenylated mRNAs are rapidly exported. Hence, to profile the transcripts at different stages of processing, polyA-isolation methods are not suited. Inspired by previous work (Morlan et al., 2012; Gregory Smaldone, personal communication), we developed a method to remove ribosomal RNA (rRNA) from rice samples, specifically using the INTACT-purified nuclei as our input. The steps of this method include isolation of total RNA from INTACT-purified nuclei, removal of residual genomic DNA, annealing of DNA probes to rRNA, degradation of rRNA with RNase specific to RNA:DNA hybrids, and degradation of the DNA probes. At the end of the protocol, the RNA sample is ready for RNA-Seq library construction.
Part I: INTACT purification of nuclei
Materials and Reagents
- Pipette tips (P20, P200 and P1000, nuclease-free, e.g., USA Scientific, catalog numbers: 1123-1710 , 1120-8780 , 1126-7510 )
- (Optional) 13 ml Falcon tube
- 30 μm cell strainer (Sysmex, CellTrics, catalog number: 04-004-2326 )
- 15 ml Falcon tubes (nuclease-free, e.g., VWR, catalog number: 89039-666 )
- 1.5 ml Eppendorf tubes (nuclease-free, e.g., Denville Scientific, catalog number: C2170 )
- Pasteur pipettes (nuclease-free, e.g., Phenix Research Products, catalog number: PP-137038C )
- Slide
- Coverslip
- PCR tube strips (nuclease-free, e.g., USA Scientific, catalog number: 1402-2708 )
- 0.22 μm syringe filters (e.g., Merck, catalog number: SLGP033RB )
- 50 ml Falcon tubes (nuclease-free, e.g., VWR, catalog number: 89039-658 )
- Aluminum foil
- Transgenic plant tissue with biotin-tagged nuclei
- INTACT binary vectors (Ron et al., 2014; Reynoso et al., 2018)
Note: They will be available at https://gateway.psb.ugent.be/search. These plasmids allow inserting a T-DNA with the promoter of your choice into the plant species of your choice. - Liquid nitrogen
- M-280 Streptavidin Dynabeads (Thermo Fisher Scientific, InvitrogenTM, catalog number: 11205D )
- NaOH
- MOPS (Sigma-Aldrich, catalog number: M1254-25G )
- Sodium chloride (NaCl) (e.g., Fisher Scientific, catalog number: S271 )
- Potassium chloride (KCl) (e.g., Fisher Scientific, catalog number: BP366-1 )
- Ethylenediaminetetraacetic acid (EDTA) (e.g., Fisher Scientific, catalog number: S311 )
- EGTA (Sigma-Aldrich, catalog number: E3889-25G )
- Spermine (Sigma-Aldrich, catalog number: S1141 )
- Spermidine (Sigma-Aldrich, catalog number: S2626 )
- Protease Inhibitor Cocktail (Sigma-Aldrich, catalog number: P9599 ) or cOmplete mini protease inhibitor tablets EDTA-free (Roche Diagnostics, catalog number: 11836170001 )
- Triton X-100 (Sigma-Aldrich, catalog number: 648466 )
- Propidium Iodide (PI) stain (Sigma-Aldrich, catalog number: P4170 )
- 10 N NaOH (see Recipes)
- 0.5 M MOPS, pH 7 (see Recipes)
- 1 M NaCl (see Recipes)
- 2 M KCl (see Recipes)
- 0.5 M EDTA (see Recipes)
- 0.5 M EGTA (see Recipes)
- 0.2 M spermine (see Recipes)
- 0.2 M spermidine (see Recipes)
- 10% Triton X-100 (see Recipes)
- Nuclei purification buffer base (see Recipes)
- NPB and NPBt buffers (see Recipes)
- Propidium Iodide (PI) stains (see Recipes)
Equipment
- Pipettes (P20, P200, P1000; e.g., Eppendorf, catalog numbers: 3123000039 , 3123000055 , 3123000063 )
- Ceramic mortars and pestles (wipe clean with RNaseZap [Thermo Fisher Scientific, InvitrogenTM, catalog number: AM9780 ] or other RNase removal product)
- Centrifuge for 15 ml Falcon tubes (e.g., Eppendorf, model: 5810 R )
- DynamagTM-15 Magnet (Thermo Fisher Scientific, model: DynamagTM-15, catalog number: 12301D )
Note: Can be replaced with a homemade version. For example, a strong rare earth (neodymium) magnet (e.g., 1 x 10 cm bar) can be taped inside a 50 ml Falcon tube and padding can be added to keep 15 ml Falcon tubes in place next to the magnet (see Figure 1).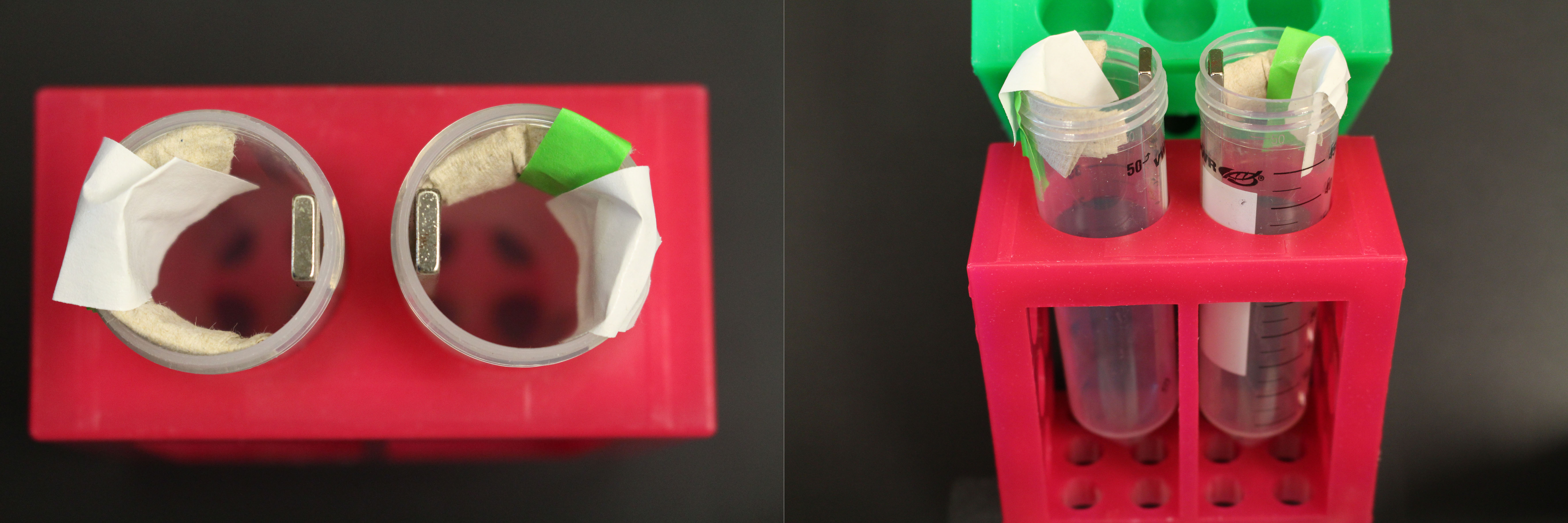
Figure 1. Homemade magnet for two 15 ml Falcon tubes. Two neodymium magnets are placed inside 50 ml Falcon tubes held in a tube rack. The attraction between the two magnets holds them in place. To prop the 15 ml tubes to correct position against the magnet, paper towel is taped inside the 50 ml tube. - DynamagTM-2 Magnet (Thermo Fisher Scientific, model: DynamagTM-2, catalog number: 12321D ), or a homemade version of this
- Hemocytometer (SCK Films, In-Cyto, catalog number: DHC-N01-2 )
- BD AdamsTM NutatorTM Single-Speed Orbital Mixer (e.g., Fisher Scientific, catalog number: 14-062)
Manufacturer: BD, catalog number: 421105/DEL . - Fridge and/or cold room at 4 °C
- Fluorescent microscope with filters to visualize either DAPI or PI stain (Edmund Optics, catalog numbers: 86-371 , 67-008 )
Procedure
文章信息
版权信息
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Reynoso, M. A., Pauluzzi, G. C., Cabanlit, S., Velasco, J., Bazin, J., Deal, R., Brady, S., Sinha, N., Bailey-Serres, J. and Kajala, K. (2018). Isolation of Nuclei in Tagged Cell Types (INTACT), RNA Extraction and Ribosomal RNA Degradation to Prepare Material for RNA-Seq. Bio-protocol 8(7): e2458. DOI: 10.21769/BioProtoc.2458.
- Reynoso, M.A, Pauluzzi, G., Kajala, K., Cabanlit, S., Velasco, J., Bazin, J., Deal, R.B., Sinha, N.R., Brady, S.M., Bailey-Serres J. (2018). Nuclear transcriptomes at high resolution using retooled INTACT. Plant Physiol 176(1): 270-281.
分类
植物科学 > 植物分子生物学 > RNA > RNA 测序
植物科学 > 植物分子生物学 > RNA > RNA 提取
分子生物学 > RNA > RNA 测序
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link