- EN - English
- CN - 中文
Cell Type-specific Metabolic Labeling of Proteins with Azidonorleucine in Drosophila
果蝇中含叠氮正亮氨酸蛋白质的细胞类型特异性代谢标记
发布: 2017年07月20日第7卷第14期 DOI: 10.21769/BioProtoc.2397 浏览次数: 8960
评审: Jyotiska ChaudhuriManish ChamoliRosario Gomez-Garcia
Abstract
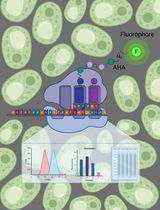
Advanced mass spectrometry technology has pushed proteomic analyses to the forefront of biological and biomedical research. Limitations of proteomic approaches now often remain with sample preparations rather than with the sensitivity of protein detection. However, deciphering proteomes and their context-dependent dynamics in subgroups of tissue-embedded cells still poses a challenge, which we meet with a detailed version of our recently established protocol for cell-selective and temporally controllable metabolic labeling of proteins in Drosophila. This method is based on targeted expression of a mutated variant of methionyl-tRNA-synthetase, MetRSL262G, which allows for charging methionine tRNAs with the non-canonical amino acid azidonorleucine (ANL) and, thus, for detectable ANL incorporation into nascent polypeptide chains.
Keywords: Metabolic labeling (代谢标记)Background
The protein composition of any given cell is intimately linked to its state of differentiation and functionality. Changes in a cell’s proteome may reflect its response to cell-intrinsic cues or to signals originating from elsewhere inside the respective organism or its environment. In turn they are indicative of the significance of those signaling cues. Deciphering proteomes and their dynamics in a cell type-specific fashion has thus become a main focus in current research, reaching a better understanding of molecular events underlying physiological or pathophysiological processes. Any proteomic approach in this direction, however, is challenged by the heterogeneity of cell types that are interconnected within a tissue or organ of interest. In the brain, for instance, different types of neurons and glial cells form the networks required to control animal or human behavior. Moreover, it is well established that information processing within these networks leading to long-term memory is strictly dependent on de novo protein synthesis and degradation. While this has been exemplified for a number of neuronal proteins (e.g., immediate early gene proteins), it is obvious that proteins that are up- or down-regulated in just a limited number of cells (or even are regulated oppositely in different groups of cells) may easily escape conventional modes of detection, where cellular proteomes are averaged across entire brain areas.
A number of labeling methods for cellular proteomes have been published in the last two decades, e.g., using isotope-coded affinity tags (Gygi et al., 1999) or isobaric tags for relative and absolute quantification (Ross et al., 2004), quantitative proteomic analysis using samples from cells grown in 14N or 15N media (Washburn et al., 2002; MacCoss et al., 2003), and stable isotope labeling by amino acids in cell culture (Ong et al., 2002; Andersen et al., 2005). Moreover puromycin (Schmidt et al., 2009) and non-canonical amino acids, e.g., azidohomoalanine (AHA) or homopropargylglycine, in combination with click chemistry have been used to decipher cellular proteomes (Link et al., 2003; Link and Tirrell, 2003; Beatty et al., 2006; Dieterich et al., 2006; Link et al., 2006; Dieterich et al., 2010). All of these strategies, however, fail to uncover cell-type specific proteomes within tissue or organ samples. Most recently, novel strategies to resolve this issue have been reported for C. elegans and Drosophila (Elliott et al., 2014; Erdmann et al., 2015; Yuet et al., 2015). They have in common the use of either a mutated aminoacyl-tRNA synthetase or an orthogonal aminoacyl-tRNA synthetase/tRNA for tagging of newly synthesized proteins with food-supplied non-canonical amino acids. Specifically, we could show that upon cell type-specific expression of a mutant Methionyl-tRNA synthetase (MetRSL262G) as achieved by employing the well-established Gal4/UAS-system, the non-canonical amino acid ANL can be incorporated into proteins of selectable cell types in living Drosophila larvae and adult flies. An accompanying study by Niehues et al. (2015) used this method to show the causal involvement of mutated glycyl-tRNA synthetase in a model for the neurodegenerative Charcot Marie Tooth disease.
ANL-containing proteins can either be analyzed in protein extracts by using biochemistry and mass spectrometry or can be visualized in situ by fluorescence microscopy (Erdmann et al., 2015; Niehues et al., 2015). For more information see ‘Click Chemistry (CuAAC) and detection of tagged de novo synthesized proteins’. The following protocol details the metabolic labeling of proteins in larvae and adult flies with ANL.
Materials and Reagents
- Fly vials (e.g., VWR, catalog number: 734-2254 )
- Fly vial plugs (e.g., Carl Roth, catalog number: PK13.1 )
- Gal4 activator strains of choice (e.g., C57-Gal4 for muscle-specific expression [from Ulrich Thomas, Magdeburg, Germany], elavC155-Gal4 for pan-neuronal expression [from Bloomington stock center, Bloomington, Indiana, USA], repo-Gal4 for glial expression [from Christian Klämbt, Münster, Germany])
- UAS-dMetRSL262G effector strains [available at request from Daniela C. Dieterich & Ulrich Thomas]. As described in Erdmann et al. (2015) various lines expressing dMetRSL262G either tagged with 3xmyc or EGFP are available
Note: We traditionally use ONM. The standard corn meal medium has also been successfully used in Niehues et al. (2015) for ANL labeling, thus, we anticipate that other media can be used as well without any limitations. - Otto-normal-medium (ONM, see Recipes)
- Agar-Agar (Carl Roth, catalog number: 5210 )
- Semolina (local food store)
- Mashed raisins (local food store)
- Baker’s yeast (local food store)
- Sugar beet sirup (local food store)
- Honey (local food store)
- Tap water
- 20% (w/v) Nipagin (see Recipes)
- 200 mM ANL stock solution (for the synthesis of ANL see [Link et al., 2007; Ngo et al., 2009; Erdmann et al., 2015]) (see Recipes)
Equipment
- Beaker (kitchen/household grade)
- Immersion blender (kitchen/household grade)
- Paintbrush (art supplies)
- Fly incubator (e.g., SANYO, model: MIR-553 )
- Hotplate (kitchen/household grade)
- Pot (kitchen/household grade)
- Tablespoon (kitchen/household grade)
Procedure
文章信息
版权信息
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Erdmann, I., Marter, K., Kobler, O., Niehues, S., Bussmann, J., Müller, A., Abele, J., Storkebaum, E., Thomas, U. and Dieterich, D. C. (2017). Cell Type-specific Metabolic Labeling of Proteins with Azidonorleucine in Drosophila. Bio-protocol 7(14): e2397. DOI: 10.21769/BioProtoc.2397.
分类
生物化学 > 蛋白质 > 标记
系统生物学 > 蛋白质组学 > 整个机体
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link