- EN - English
- CN - 中文
Formation of Minimised Hairpin Template-transcribing Dumbbell Vectors for Small RNA Expression
用于小分子RNA表达的最小化发夹模板转录哑铃形载体生成
发布: 2017年06月05日第7卷第11期 DOI: 10.21769/BioProtoc.2313 浏览次数: 10273
评审: Longping Victor TseGal HaimovichAnonymous reviewer(s)
Abstract
A major barrier for using non-viral vectors for gene therapy is the short duration of transgene expression in postmitotic tissues. Previous studies showed transgene expression from conventional plasmid fell to sub-therapeutic level shortly after delivery even though the vector DNA was retained, suggesting transcription was silenced in vivo (Nicol et al., 2002; Chen et al., 2004). Emerging evidence indicates that plasmid bacterial backbone sequences are responsible for the transcriptional repression and this process is independent of CpG methylation (Chen et al., 2008). Dumbbell-shaped DNA vectors consisting solely of essential elements for transgene expression have been developed to circumvent these drawbacks. This novel non-viral vector has been shown to improve transgene expression in vitro and in vivo (Schakowski et al., 2001 and 2007). Here we describe a novel method for fast and efficient production of minimised small RNA-expressing dumbbell vectors. In brief, the PCR-amplified promoter sequence is ligated to a chemically synthesized hairpin RNA coding DNA template to form the covalently closed dumbbell vector. This new technique may facilitate applications of dumbbell-shaped vectors for preclinical investigation and human gene therapy.
Keywords: Dumbbell vector (哑铃形载体)Background
With regard to delivery, a small vector size is advantageous improving extracellular transport including extravasation and diffusion through the extracellular matrix network as well as cellular uptake and nuclear diffusion. Various methods for dumbbell vector production have been developed over the time including methods for the generation of dumbbells expressing small RNAs such as small hairpin RNAs (shRNAs) and microRNAs (miRNAs) (Schakowski et al., 2001; Taki et al. 2004). These vectors usually harbour redundant sequences as the expressed RNAs are self-complementary. We eliminated redundant sequences generating minimised dumbbell vectors in which transcription goes around the hairpin structure of the dumbbell itself (Jiang et al., 2016). Such minimised dumbbell vectors can be as short as 130 bp representing the smallest expression vectors ever reported. An illustrated comparison between a conventional plasmid, a dumbbell harbouring a linear expression cassette, and a novel hairpin template-transcribing dumbbell vector is shown in Figure 1. This novel protocol facilitates the production of the new minimised small RNA expression dumbbell vectors.
.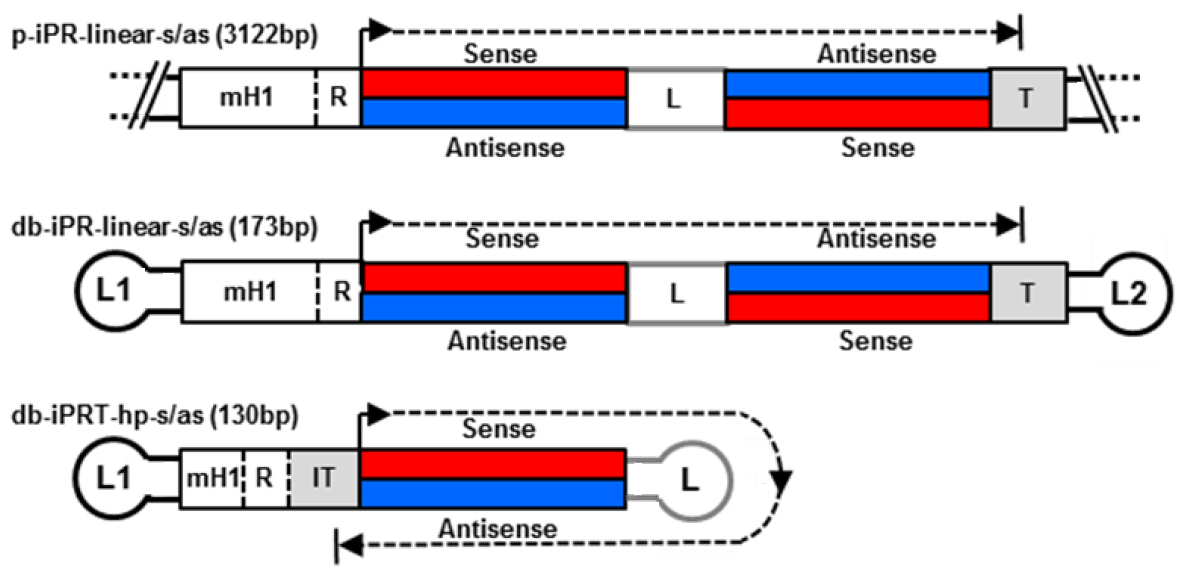
Figure 1. Structures of small hairpin RNA-expressing plasmid and dumbbell vectors. Upper two: conventional plasmid p-iPR-linear-s/as and dumbbell db-iPR-linear-s/as vectors with linear shRNA expression cassettes and integrated promoter-restriction endonuclease site element (iPR). Lower vector: minimized hairpin template (hp) dumbbell harboring an iPRT element. R indicates a restriction overhang ligation site. T indicates termination signal. IT indicates inverted termination signal. Loops L1 and L2 are (T)4 tetra loops.
Materials and Reagents
- 0.2-10 μl pipette tips, Corning® Isotip® filtered (Corning, catalog number: 4807 )
- 1-200 μl pipette tips (Corning, Axyge®, catalog number: TF-200-R-S )
- 100-1,000 μl pipette tips (Corning, Axygen®, catalog number: TF-1000-R-S )
- 1.5 ml microcentrifuge tubes (RNase, DNase and Pyrogen-Free) (Corning, Axygen®, catalog number: MCT-150-C )
- 0.2 ml thin-walled PCR tubes (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 3412 )
- Falcon® 50 ml conical centrifuge tubes (Corning, Falcon®, catalog number: 352070 )
- pSuper-basic vector (Oligoengine, catalog number: VEC-pBS-0002 )
- miRNA-coding oligonucleotide (PAGE purified) 5’-pGATCTAGCACGACTCGCAGCTCCCAAGAGCCTAACCCGTGGATTTAAACGGTAAACATCACAAGTTAGGGTCTCAGGGACTGAGAGGAGCGCAA-3’
- Oligonucleotides for minimal H1 (mH1) promoter (PAGE purified)
mH1-Fw: 5’-pAATTCATATTTGCATGTCGCTATGTGTTCTGGGAAATCACCATAAACGTGAAATGTCTTTGGATTTGGGAATCTTATAAGTTCTGTATGAGAGCACAGA-3’
mH1-Rv: 5’-pGATCTCTGTGCTCTCATACAGAACTTATAAGATTCCCAAATCCAAAGACATTTCACGTTTATGGTGATTTCCCAGAACACATAGCGACATGCAAATATG-3’ - UltraPureTM DNase/RNase-Free distilled water (Thermo Fisher Scientific, InvitrogenTM, catalog number: 10977015 )
- Magnesium chloride hexahydrate (MgCl2·6H2O) (Sigma-Aldrich, catalog number: M2670-100G )
- dNTP set 100 mM solutions (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: R0181 )
- Oligonucleotide primers for mH1 promoter amplification (HPCL purified)
NbBpu-Fw
5’-pTTAGGAGTTTTCTCCTAAGCATATTTGCATGTCGCTATGTGTTCTG-3’
BamHI-Rv
5’-TGCAGGATCCCTGTGCTCTCATACAGAACTTATAAGATTCCC-3’ - Taq DNA polymerase, recombinant (5 U/µl) (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: EP0402 )
- QIAquick PCR Purification Kit (QIAGEN, catalog number: 28106 )
- 10x FD buffer (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: B64 )
- Nb.Bpu10I (5 U/µl) (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: ER1681 )
- FastDigest BamHI (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: FD0054 )
- shRNA-coding oligonucleotide (PAGE purified) 5’-pGATCTAAAAAGAGCTGTTTCTGAGGAGCCTCTCTTGAAGGCTCCTCAGAAACAGCTCTTTTTA-3’
- T4 DNA ligase (5 U/µl) (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: EL0014 )
- Neutralizing oligonucleotide (HPLC purified)
5’-TTAGGAGTTTTCTCCTAA-3’ - Adenosine 5’-triphosphate disodium salt hydrate (Sigma-Aldrich, catalog number: A2383-1G )
- FastDigest BglII (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: FD0083 )
- T7 DNA polymerase (10 U/µl) (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: EP0081 )
- Phenol solution (Sigma-Aldrich, catalog number: P4557-100ML )
- Chloroform (Sigma-Aldrich, catalog number: 288306-1L )
- 3-methyl-1-butanol (Sigma-Aldrich, catalog number: 309435-100ML )
- Ethanol, absolute (Fisher Scientific, catalog number: BP28184 )
- 3 M potassium acetate (pH 4.8)
- Sodium acetate (Sigma-Aldrich, catalog number: S2889-250G )
- FastDigest EcoRI (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: FD0274 )
- Agarose, LE, analytical grade (Promega, catalog number: V3125 )
- Ethidium bromide solution (Bio-Rad Laboratories, catalog number: 1610433 )
- GeneRuler DNA ladder mix (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: SM0331 )
- Sodium chloride (NaCl) (Sigma-Aldrich, catalog number: S9888-500G )
- Tris-HCl (Powder) (Roche Diagnostics, catalog number: 10812846001 )
- EDTA (Sigma-Aldrich, catalog number: EDS-100G )
- 10x hybridization buffer (see Recipes)
- TE buffer (see Recipes)
Equipment
- Pipettes (Gilson, PIPETMAN® Classic, models: P2, P20N, P200N, and P1000N )
- Standard thermal cycler (Thermo Fisher Scientific, Applied BiosystemsTM, model: GeneAmp PCR System 9700 )
Note: This product has been discontinued. - Gel doc (Bio-Rad Gel Doc Imager)
- Gel running apparatus (Thermo Fisher Scientific, Amersham BiosciencesTM)
- Gel staining tray
- Benchtop centrifuge (Eppendorf, model: 5430 R )
- Heat block (Eppendorf, model: Thermomixer® Comfort )
- Spectrophotometer (Thermo Fisher Scientific, Thermo ScientificTM, model: NanoDrop 2000 )
- Glass beaker (Schott, Duran)
- Microwave (Panasonic)
Software
- ImageJ (http://imagej.nih.gov/ij/)
Procedure
文章信息
版权信息
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Jiang, X. and Patzel, V. (2017). Formation of Minimised Hairpin Template-transcribing Dumbbell Vectors for Small RNA Expression. Bio-protocol 7(11): e2313. DOI: 10.21769/BioProtoc.2313.
分类
分子生物学 > DNA > DNA 克隆
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link