- EN - English
- CN - 中文
Ultradeep Pyrosequencing of Hepatitis C Virus to Define Evolutionary Phenotypes
超深度焦磷酸测序鉴定丙型肝炎病毒的进化表型
发布: 2017年05月20日第7卷第10期 DOI: 10.21769/BioProtoc.2284 浏览次数: 6846
评审: Yannick DebingRaju GhoshVamseedhar RayaproluAnonymous reviewer(s)
Abstract
Analysis of hypervariable regions (HVR) using pyrosequencing techniques is hampered by the ability of error correction algorithms to account for the heterogeneity of the variants present. Analysis of between-sample fluctuations to virome sub-populations, and detection of low frequency variants, are unreliable through the application of arbitrary frequency cut offs. Cumulatively this leads to an underestimation of genetic diversity. In the following technique we describe the analysis of Hepatitis C virus (HCV) HVR1 which includes the E1/E2 glycoprotein gene junction. This procedure describes the evolution of HCV in a treatment naïve environment, from 10 samples collected over 10 years, using ultradeep pyrosequencing (UDPS) performed on the Roche GS FLX titanium platform (Palmer et al., 2014). Initial clonal analysis of serum samples was used to inform downstream error correction algorithms that allowed for a greater sequence depth to be reached. PCR amplification of this region has been tested for HCV genotypes 1, 2, 3 and 4.
Keywords: Ultradeep pyrosequencing (超深度焦磷酸测序)Background
Analysis of UDPS datasets derived from virus amplicons frequently relies on software tools that are not optimized for amplicon analysis, assume random incorporation of sequencing mutations and are focused on finding true sequences rather than false variants. These difficulties are further complicated by the presence of hypervariable regions present in RNA virus genomes. Many studies utilizing UDPS look to overcome these issues by applying arbitrary frequency cut offs to the data, resulting in the loss of minor variants. Here, a temporally matched clonal dataset, together with an error correction methodology designed to overcome the problems outlined, facilitated the retention of valuable sequence information.
Materials and Reagents
- 1.5 ml tube (SARSTEDT, catalog number: 72.690.001 )
- 200 µl MicroAmp® PCR tube (Thermo Fisher Scientific, Applied BiosystemsTM, catalog number: N8010840 )
- Clean stainless steel blade
- One Shot® TOP10 Competent Cells (Thermo Fisher Scientific, InvitrogenTM, catalog number: C404003 )
- QIAamp® Viral RNA mini kit (QIAGEN, catalog number: 52904 )
- Random primer (Promega, catalog number: C1181 )
- Deoxynucleoside triphosphate (dNTP’s, 100 mM) set, PCR grade (Roche Molecular Systems, catalog number: 11969064001 )
- AMV reverse transcriptase (Promega, catalog number: M5101 )
- RNasin® Ribonuclease inhibitor (Promega, catalog number: N2511 )
- Outer-forward primer: 5’- ATGGCATGGGATATGAT -3’ (10 pmol/µl, Eurofins)
- Outer-reverse primer: 5’- AAGGCCGTCCTGTTGA -3’ (10 pmol/µl, Eurofins)
- Inner-forward primer: 5’- GCATGGGATATGATGATGAA -3’ (10 pmol/µl, Eurofins)
- Inner-reverse primer: 5’- GTCCTGTTGATGTGCCA -3’ (10 pmol/µl, Eurofins)
- Pwo DNA polymerase (5 U/µl,) including 10x reaction buffer (- MgSO4) and MgSO4 stock solution (25 mM) (Roche Molecular Systems, catalog number: 11644955001 )
- dH2O (Sigma-Aldrich, catalog number: W4502 )
- Sybr safe DNA gel stain (Thermo Fisher Scientific, InvitrogenTM, catalog number: S33102 )
- Agarose (Sigma-Aldrich, catalog number: A9539 )
- GeneRuler 100 bp Plus DNA ladder (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: SM0323 )
- Gel extraction kit (QIAGEN, catalog number: 28704 )
- CloneJet PCR Cloning Kit (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: K1231 )
- GeneJet Plasmid Miniprep Kit (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: K0503 )
- Trizma® base (Sigma-Aldrich, catalog number: T1503 )
- Acetic acid glacial (BDH Laboratory Supplies, catalog number: 10001CU )
- Ethylenediaminetetraacetic acid solution 0.5 M (EDTA) (Sigma-Aldrich, catalog number: 03690 )
- 1x TAE (see Recipes)
Equipment
- PCR thermal cycler (Thermo Fisher Scientific, Applied BiosystemsTM, model: Applied Biosystems® 2720 )
- BioPhotometer (Eppendorf, http://arboretum.harvard.edu/wp-content/uploads/Biophotometer-manual.pdf)
- Water bath (JULABO, model: SW22 )
- Orbital shaker incubator (Grant, model: ES-80 )
- Ultraviolet transilluminator (UVP, model: TMW-20 )
Software
- SFFFile tools (Roche Molecular Systems)
- k-mer error correction (KEC) and empirical threshold (ET) (Skums et al., 2012)
- MEGA 6.0 (Tamura et al., 2013)
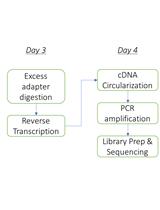
Procedure
文章信息
版权信息
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Palmer, B. A., Dimitrova, Z., Skums, P., Crosbie, O., Kenny-Walsh, E. and Fanning, L. J. (2017). Ultradeep Pyrosequencing of Hepatitis C Virus to Define Evolutionary Phenotypes. Bio-protocol 7(10): e2284. DOI: 10.21769/BioProtoc.2284.
分类
分子生物学 > RNA > RNA 测序
微生物学 > 微生物遗传学 > RNA > 测序
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link