- EN - English
- CN - 中文
Evaluation of Plasmid Stability by Negative Selection in Gram-negative Bacteria
通过革兰氏阴性细菌的负向选择评估质粒稳定性
发布: 2017年05月05日第7卷第9期 DOI: 10.21769/BioProtoc.2261 浏览次数: 12872
评审: Valentine V TrotterAlba BlesaSeda Ekici

相关实验方案
从不同难降解细菌中高质量分离质粒的方案:农杆菌、根瘤菌和苏云金芽孢杆菌
Preshobha Kodackattumannil [...] Khaled M. A. Amiri
2023年08月05日 2524 阅读
Abstract
Plasmid stability can be measured using antibiotic-resistance plasmid derivatives by positive selection. However, highly stable plasmids are below the sensitivity range of these assays. To solve this problem we describe a novel, highly sensitive method to measure plasmid stability based on the selection of plasmid-free cells following elimination of plasmid-containing cells. The assay proposed here is based on an aph-parE cassette. When synthesized in the cell, the ParE toxin induces cell death. ParE synthesis is controlled by a rhamnose-inducible promoter. When bacteria carrying the aph-parE module are grown in media containing rhamnose as the only carbon source, ParE is synthesized and plasmid-containing cells are eliminated. Kanamycin resistance (aph) is further used to confirm the absence of the plasmid in rhamnose grown bacteria.
Keywords: Plasmid stability (质粒稳定性)Background
lassically, plasmid stability has been measured by positive selection using antibiotic-resistance plasmid derivatives. Cells harbouring the studied plasmid are positively selected in the presence of the selection antibiotic (Gerdes et al., 1985; del Solar et al., 1987). The main drawback of this technique is its sensitivity; highly stable plasmids are below the sensitivity of these assays. To solve this problem alternative methods relying on the direct selection of plasmid-free cells such as the tetAR-chlortetracycline system, have been described (Bochner et al., 1980; Maloy and Nunn, 1981; Garcia-Quintanilla et al., 2006). Limitations of the tetAR-chlortetracycline method include poor reproducibility and the frequent occurrence of false positives (Li et al., 2013). Here, we describe a novel, highly sensitive plasmid stability assay based on the counter-selection of plasmid-containing cells. This assay is based on a cassette containing a ParE toxin-encoding gene controlled by a rhamnose-inducible promoter and a kanamycin resistance gene (aph) (Figure 1) (Maisonneuve et al., 2011). ParE is the toxin of the toxin-antitoxin system parDE, and targets the DNA gyrase, blocks DNA replication and induces DNA breaks leading to cell death (Jiang et al., 2002). The aph-parE cassette is inserted into the plasmid of interest using homologous recombination. Upon induction of PparE in minimal media containing rhamnose as the only carbon source, only plasmid-free cells survive (Lobato-Marquez et al., 2016). Kanamycin is then used to confirm the loss of the plasmid (Figure 2).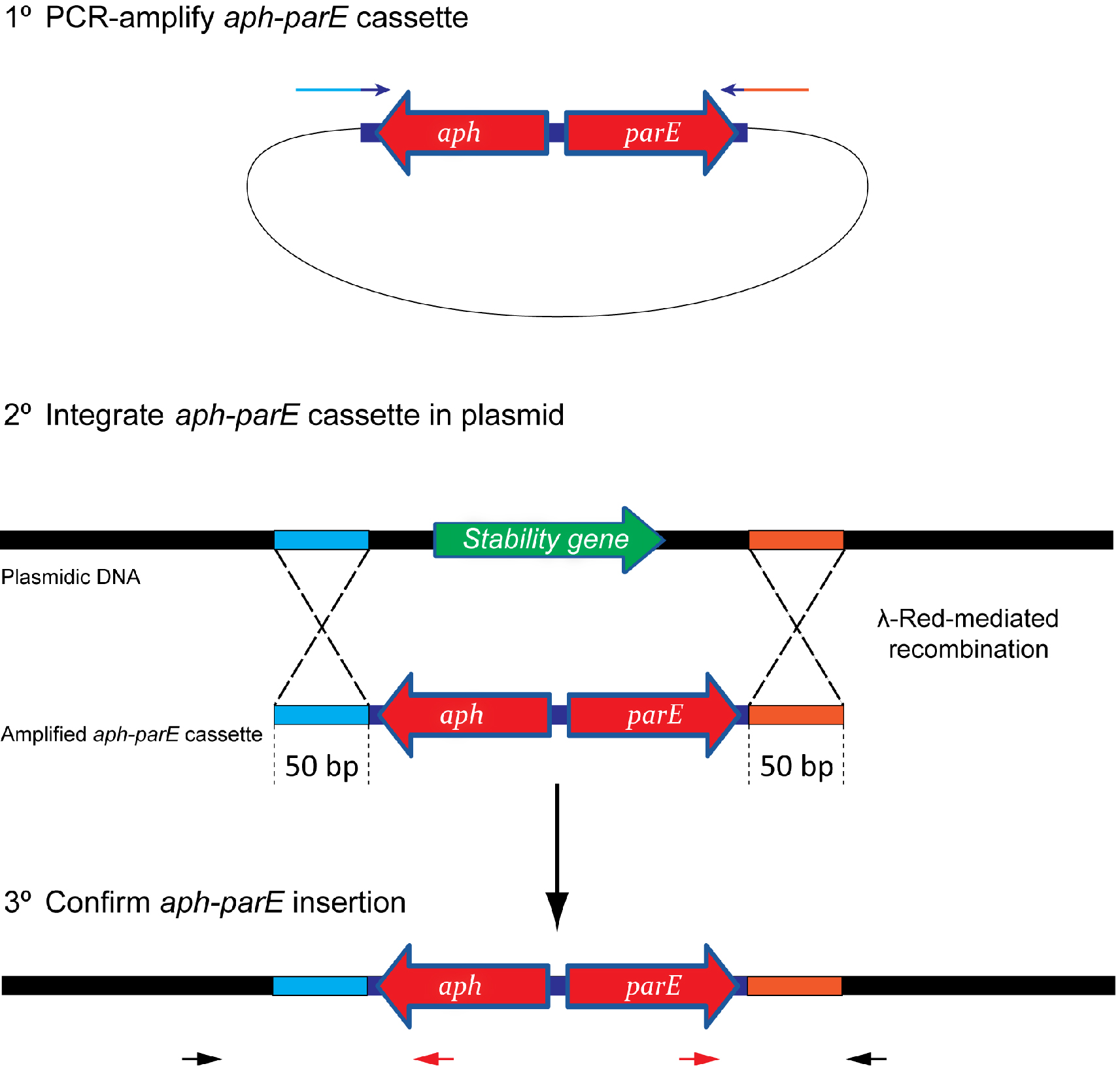
Figure 1. Scheme showing the integration process of the aph-parE cassette into the plasmid of interest. (1) The aph-parE cassette is first amplified by PCR using pKD267 plasmid as template. (2) Then, cells harbouring a plasmid encoding λ-Red recombinase are electroporated with aph-parE DNA fragment. λ-Red recombinase directs the specific integration of the aph-parE module into the plasmid region containing the 50 bp upstream and 50 bp downstream homologous sequences included in the oligos used for PCR. (3) Confirm the aph-parE insertion by using primers annealing with the cassette (red arrows) and with the plasmid (black arrows).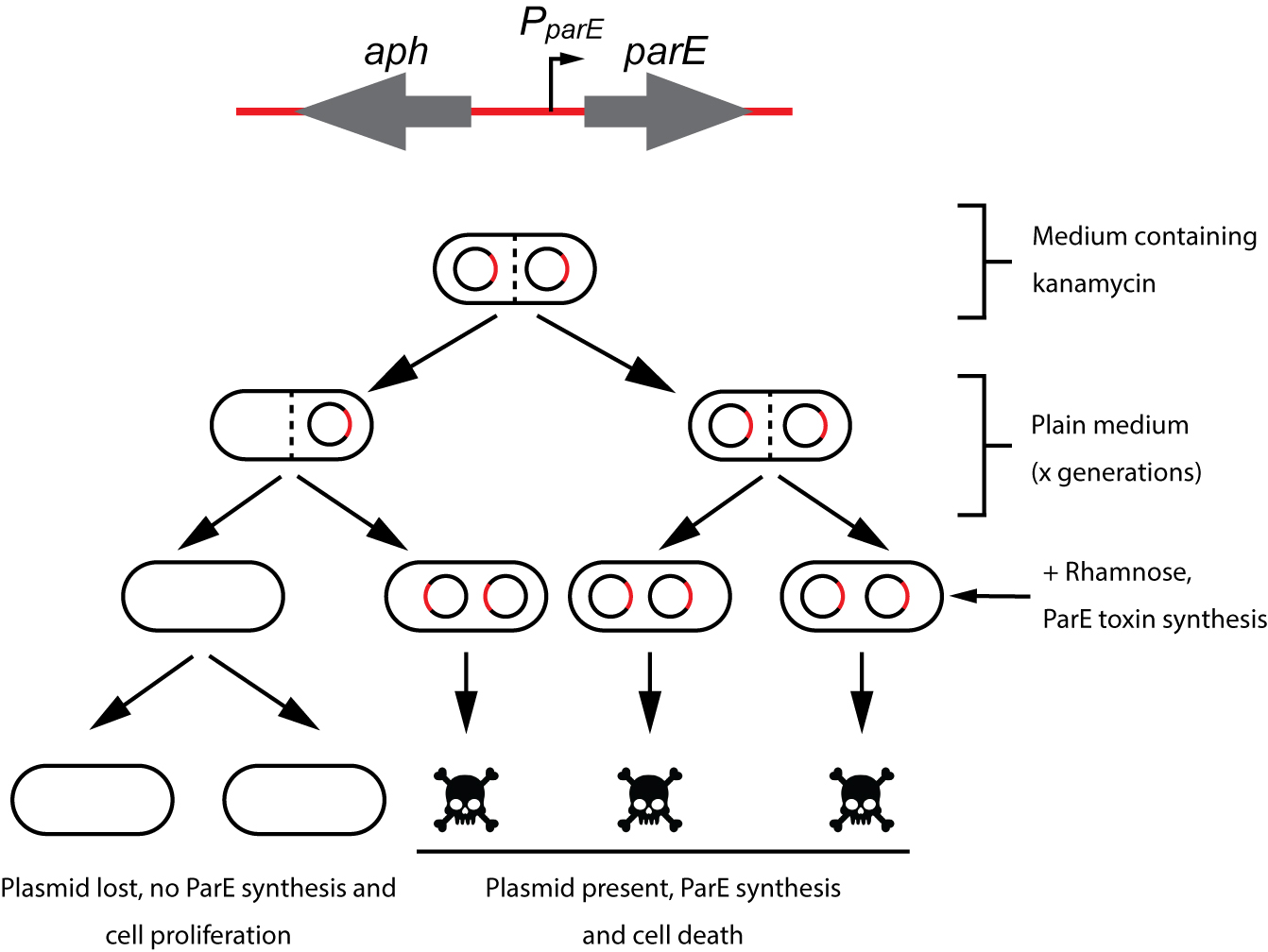
Figure 2. Plasmid stability procedure. To avoid plasmid loss, the strain carrying the aph-parE cassette is initially grown under antibiotic selection pressure (using kanamycin). When kanamycin is removed from the medium, the plasmid of interest will be lost after a certain number of generations. Plasmid-free cells are selected when the culture is plated in M9-rhamnose plates containing rhamnose as the only carbon source. Modified from Lobato-Marquez et al., 2016.
Materials and Reagents
- Pipette tips
- 1.5 ml Eppendorf tubes
- Millipore 0.22 µm pore size filter (EMD Millipore, catalog number: SLGP033RS )
- 9-cm sterile Petri dishes
- Electroporation cuvettes 0.2 cm gap (Bio-Rad Laboratories, catalog number: 1652082 )
- Glass beads for bacterial plating (VWR, catalog number: 201-0279 )
- pKD267 plasmid (described in Maisonneuve et al., 2011)
- pKD46 plasmid (described in Datsenko and Wanner, 2000)
- Expand High Fidelity DNA polymerase (Roche Molecular Systems, catalog number: 11732641001 )
- DpnI restriction enzyme (New England Biolabs, catalog number: R0176 )
- PCR purification kit (5Prime, catalog number: 2300610 )
- Plasmid purification kit (Roche Molecular Systems, catalog number: 11754777001 )
- 4 °C chilled sterile distilled MilliQ water
- Distilled MilliQ water
- Ampicillin sodium salt (Sigma-Aldrich, catalog number: A0166 )
- L-arabinose (Sigma-Aldrich, catalog number: A3256 )
- Kanamycin (Sigma-Aldrich, catalog number: K1876 )
- D-glucose (Sigma-Aldrich, catalog number: G8270 )
- Bacto tryptone (BD, BactoTM, catalog number: 211705 )
- Bacto yeast extract (BD, BactoTM, catalog number: 288620 )
- Sodium chloride (NaCl) (VWR, BDH®, catalog number: 102415K )
- European bacteriological agar (Conda, catalog number: 1800 )
- Sodium phosphate dibasic (Na2HPO4) (Sigma-Aldrich, catalog number: 71645 )
- Potassium phosphate monobasic (KH2PO4) (Sigma-Aldrich, catalog number: P9791 )
- Ammonium chloride (NH4Cl) (Sigma-Aldrich, catalog number: A9434 )
- Calcium chloride dihydrate (CaCl2·2H2O) (Sigma-Aldrich, catalog number: C3881 )
- Magnesium sulfate (MgSO4) (VWR, BDH®, catalog number: 291175X )
- Vitamin B1 (Thiamine) (Sigma-Aldrich, catalog number: T4625 )
- L-rhamnose monohydrate (Sigma-Aldrich, catalog number: R3875 )
- Potassium chloride (KCl)
- Na2HPO4·12H2O
- Glycerol (v/v) (Sigma-Aldrich, catalog number: G5516 )
- Luria-Bertani (LB) broth medium (see Recipes)
- Luria Bertani plates (see Recipes)
- M9 (10x) (see Recipes)
- CaCl2/MgSO4 solution (100x)
- M9 minimum medium (see Recipes)
- M9-rhamnose plates (see Recipes)
- Phosphate saline buffered (PBS) (see Recipes)
- 10% sterile glycerol (see Recipes)
- L-rhamnose (see Recipes)
Equipment
- Selection of single channel pipettes (2 µl, 20 µl, 20 µl, 1,000 µl) (Gilson, P-2, P-20, P-200, P-1000)
- Glass 100 ml flasks (Fisher Scientific, Fisherbrand)
- Glass 50 ml flasks (Fisher Scientific, Fisherbrand)
- MiniSpin® Eppendorf benchtop centrifuge (Eppendorf, model: MiniSpin® )
- Bacterial shaking incubator (Eppendorf, New Brunswick, model: Innova® 4000 )
- MicroPulserTM electroporator (Bio-Rad Laboratories, model: MicroPulser Electroporator , catalog number: 1652100)
- Eppendorf refrigerated centrifuge (Eppendorf, model: 5804 )
- Spectrophotometer (Thermo Fisher Scientific, Thermo ScientificTM, model: SPECTRONICTM 200 )
- Milli-Q® integral water purification system for ultrapure (Nanopure) water
- DNA SpeedVac (Savant Systems, model: SpeedVac DNA 110 )
- Autoclave (Prestige Medical, catalog number: 210004 )
Software
- GraphPad Prism Software (https://www.graphpad.com/scientific-software/prism/)
Procedure
文章信息
版权信息
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Lobato-Márquez, D. and Molina-García, L. (2017). Evaluation of Plasmid Stability by Negative Selection in Gram-negative Bacteria. Bio-protocol 7(9): e2261. DOI: 10.21769/BioProtoc.2261.
分类
微生物学 > 微生物遗传学 > 质粒
分子生物学 > DNA > 染色体外DNA
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link