- EN - English
- CN - 中文
Lentiviral shRNA Screen to Identify Epithelial Integrity Regulating Genes in MCF10A 3D Culture
利用慢病毒shRNA筛选鉴定调节上皮细胞MCF10A 3D完整性的基因
发布: 2016年12月05日第6卷第23期 DOI: 10.21769/BioProtoc.2050 浏览次数: 12130
评审: HongLok LungSteve JeanAnonymous reviewer(s)
Abstract
MCF10A 3D culture system provides a reductionist model of glandular mammary epithelium which is widely used to study development of glandular architecture, the role of cell polarity and epithelial integrity in control of epithelial cell functions, and mechanisms of breast cancer. Here we describe how to use shRNA screening approach to identify critical cell pathways that couple epithelial structure to individual cell based responses such as cell cycle exit and apoptosis. These studies will help to interrogate genetic changes critical for early breast tumorigenesis. The protocol describes a library of lentiviral shRNA constructs designed to target epithelial integrity and a highly efficient method for lentiviral transduction of suspension MCF10A cultures. Furthermore, protocols are provided for setting up MCF10A 3D cultures in Matrigel for morphometric and cellular response studies via structured illumination and confocal microscopy analysis of immunostained 3D structures.
Keywords: 3D cultures (三维培养)Background
All epithelial cells form highly organized tissue structures, which provide physical support and a structured scaffold for coordinated cell signaling. Such coordinated signaling across the epithelial structures is fundamental for epithelial biology; enabling dynamic joint actions of epithelial cells in regulation of organ size, shape, function and individual cell based responses (Roignot et al., 2013; Shamir and Ewald, 2014). Joint command of epithelial signaling also presents a powerful tumor suppressor mechanism by gatekeeping extrinsic and intrinsic mitogenic signals to quiescent epithelial tissues (Partanen et al., 2013; Rejon et al., 2016). However, very little is still known about genetic mechanisms coupling the status of epithelial structure with individual epithelial cell functions. MCF10A 3D Matrigel culture system is a well-established genetically tractable model of mammary epithelial architecture that is widely used to explore epithelial context-dependent cell functions (Debnath and Brugge, 2005). However, individual structures in MCF10A 3D cultures are not fully uniform in size or symmetry, which makes high-throughput screens with shRNA or cDNA reagents challenging in this system. Here, we describe protocols that expand the use of MCF10A 3D culture system from single gene studies to cell pathway level perturbation studies. The protocols for medium-throughput 3D screen using validated lentiviral shRNAs were originally used in a screen designed to identify genes with epithelial integrity-linked proliferation functions (Marques et al., [2016], screen outlined in Figure 1). However, these protocols are suitable for any reverse genetic MCF10A 3D culture study within a range of about 50 perturbed genes of interest.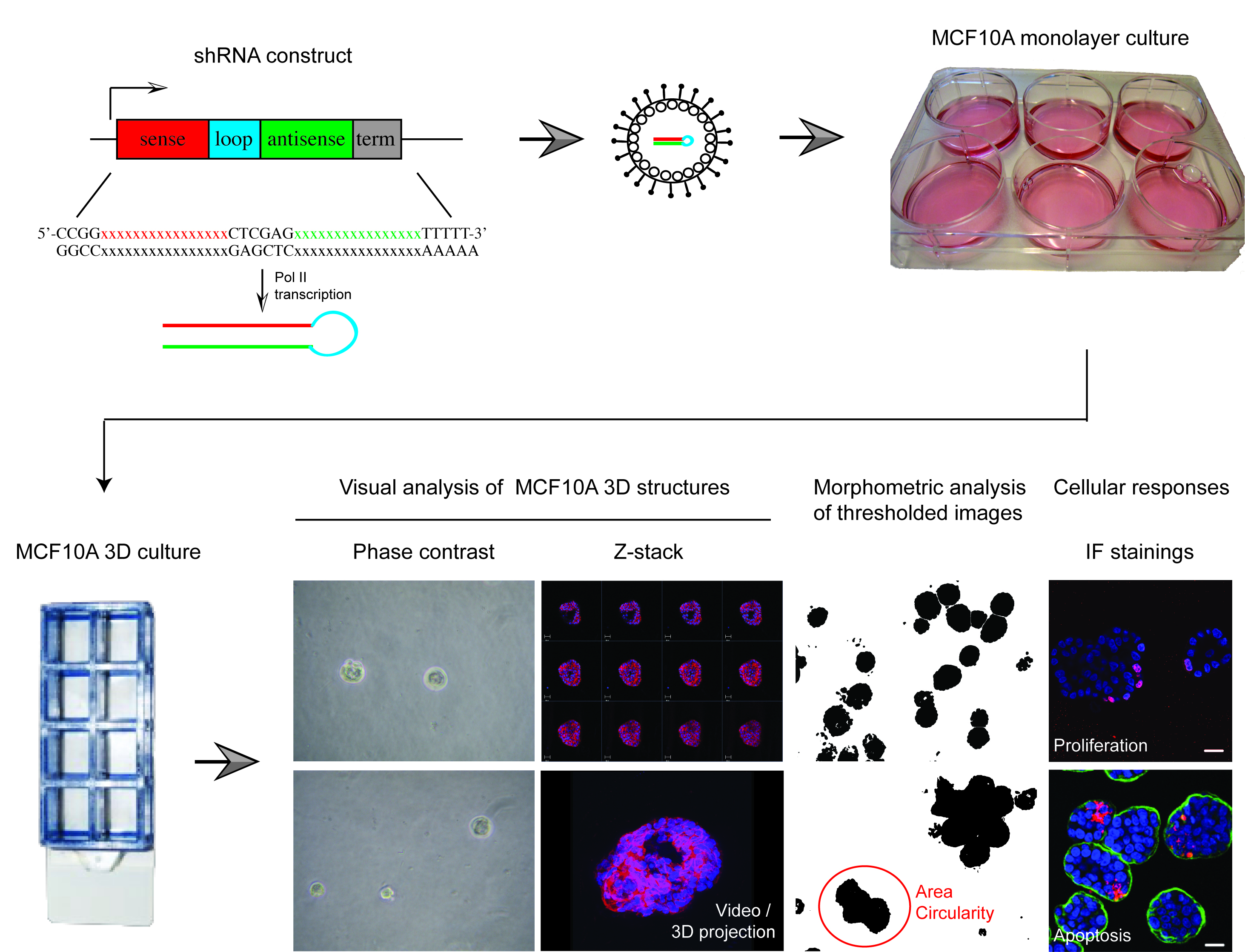
Figure 1. Overview of shRNA screen in MCF10A 3D culture designed to identify epithelial integrity regulating genes. The protocols described here were recently applied in a shRNA screen using 52 knockdown validated shRNAs, which were lentivirally transferred to MCF10A cells containing a switchable oncogenic form of Myc (MycERTM). This set up allowed two separate primary morphometric screens in MCF10A 3D cultures; one with and another without Myc oncogene challenge. These screens produced morphometric data from > 5,000 structures. The most interesting knockdown phenotypes were further analyzed with cell response markers (proliferation [i.e., Ki67], apoptosis [i.e., active caspase 3] and polarity change [i.e., α6-integrin, GM130]) and via 3D structures reconstructions obtained with confocal microscopy. The results from this screen for morphometric results have been published in Marques et al. (2016).
Materials and Reagents
- 24-well plates (VWR, catalog number: 391-3370 )
- Minisart filters 0.45 μm pore size (Sartorius, catalog number: 16537 )
- 6-well plates (VWR, catalog number: 700-1425 )
- 8-chamber slides (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 177402 )
- Microscope coverslips (MEDI PLAST FENNO, catalog number: 702-1051204 )
- Pipet tips
- 500 ml bottle
- Ultra adherent cell culture plates for 293ft cells
- Nunclon surface 10 cm (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 150350 )
- Nunclon surface 6 well plates (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 145380 )
- 293ft cells (Thermo Fisher Scientific, InvitrogenTM, catalog number: R70007 )
- MCF10A cells (ATCC, catalog number: CRL-10317TM )
- MCF10A-MycER cells (Nieminen et al., 2007)
- Plasmid and vectors:
- pCMV-dR8.91 (Delta 8.9) packaging plasmid (Marques et al., 2016)
- pCMV-VSVg envelope construct (Marques et al., 2016)
- pDSL_UGIH lentiviral shRNA vector (Alliance for Cellular Signaling) (Marques et al., 2016)
- pLKO lentiviral vector (Broad Institute TRC library, MISSION TRC-Hs 1.0 library) (Sigma-Aldrich) (Marques et al., 2016)
- pGIPZ mir-30 based vector (Open Biosystems) (Marques et al., 2016)
- Non-targeting shRNA control constructs (in pDSL_UGIH, pLKO.1 and pGIPZ lentiviral backbone) (Marques et al., 2016)
- Modified entry vector pENTR-H1 (Nieminen et al., 2007; Marques et al., 2016)
- Cell culture and transfection
- Dulbecco’s modified Eagle medium (DMEM) (Thermo Fisher Scientific, GibcoTM, catalog number: 21041-025 )
- Mammary epithelial basal medium (MCDB 170) (BioConcept AG Costumer made product based on USB powder formulation [Labome, catalog number: M2162 ] without L-glutamine; catalog number: 9-02S77-I)
- Transfection reagent - jetPEI (Polyplus, catalog number: 101-01N )
- Supplements for DMEM media (added before use; See Recipes):
- Fetal calf serum (FCS) (Biowest, catalog number: S1810-500 )
- L-glutamine (Lanza, catalog number: 17-605E )
- Penicillin-streptomycin (Lanza, catalog number: 17-602E )
- Phosphate buffered saline (PBS) (Sigma-Aldrich, catalog number: P4417 ) (Bionordika, catalog number: 17-516F/12 )
- Polybrene (Sigma-Aldrich, catalog number: H9268 )
- Matrigel® (Corning, catalog number: 356230 )
- RNeasy Mini Kit (Qiagen, catalog number: 74104 )
- ELB Lysis Buffer reagents (see Recipes)
- HEPES (Sigma-Aldrich, catalog number: H-3375 )
- EDTA (Sigma-Aldrich, catalog number: E5134 )
- Nonidet® P40 substitute (NP-40) (Sigma-Aldrich, catalog number: 74385 )
- Protease inhibitor cocktails
- Complete Mini (Roche Diagnostics, catalog number: 04693159001 )
- PhosphoStop (Roche Diagnostics, catalog number: 04906837001 )
- 0.05% trypsin (diluted from 5% trypsin-EDTA [10x]) (Thermo Fisher Scientific, GibcoTM, catalog number: 15400-054 )
- 4% paraformaldehyde phosphate buffer stock solution (PFA) (used as diluted 2%) (from powder: Sigma-Aldrich, catalog number: 16005-1Kg-R or from 20% aqueous solution Electron Microscopy Sciences, catalog number: 15713-S )
- Triton X-100, used as 0.25% dilution in PBS (Sigma-Aldrich, catalog number: 9002-93-1 )
- Blocking solution (see Recipes)
- Immunofluorescence (IF) buffer reagents (see Recipes)
- Sodium azide (NaN3) (Sigma-Aldrich, catalog number: S-2002 )
- Bovine serum albumin (BSA) (Biowest, catalog number: P6154 )
- Tween 20 (Sigma-Aldrich, catalog number: P9416 )
- Normal goat serum, used in 10% concentration (Thermo Fisher Scientific, GibcoTM, catalog number: PCN5000 )
- Counterstaining and Antibodies:
- Hoechst 33258 (Sigma-Aldrich, catalog number: 861405 )
Note: This product has been discontinued and it has been replaced by bisBenzimide H 33258 (Sigma-Aldrich, catalog number: B2883 ). - β-catenin (BD, catalog number: 610153 )
- GM-130 (BD, catalog number: 610823 )
- Anti-CD49f/α6-integrin (EMD Millipore, catalog number: CBL458 )
- E-cadherin (BD, catalog number: 610182 )
- Ki-67 (Leica Biosystems, catalog number: NCL-Ki67p )
- Active caspase-3 (Cell Signaling Technology, catalog number: 9661L )
- ZO-1 (Abcam, catalog number: ab59720 )
- Desmoplakin I+II (Abcam , catalog number: ab16434 or ab71690 )
- Desmoglein 2 (Abcam, catalog number: ab14415 )
- Power SYBR Green Cells-to-CT Kit (Thermo Fisher Scientific, AmbionTM, catalog number: 4402953 )
- Immu-MountTM reagent (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 9990412 )
- Sodium chloride (NaCl) (Sigma-Aldrich, catalog number: 31434-5KG-R )
- Supplements for MCDB 170 media (added before use; See Recipes):
- Bovine pituitary extract (BPE) (Sigma-Aldrich, catalog number: P-1476 ) when not available use BPE (150 mg/batch) from Upstate (Sigma-Aldrich, catalog number: 02-104 )
- Epithelial growth factor (EGF) (Sigma-Aldrich, catalog number: E-9644 )
- Transferrin (Sigma-Aldrich, catalog number: T-2252 )
- Isoproterenol (Sigma-Aldrich, catalog number: I-5627 )
- Hydrocortisone (Sigma-Aldrich, catalog number: H-4001 )
- Insulin (Sigma-Aldrich, catalog number: I-9278 )
- Amphotericin B (Sigma-Aldrich, catalog number: A-2942 )
- Gentamicin (Sigma-Aldrich, catalog number: G-1397 )
- 100 nM 4-hydroxytamoxifen used to activate MycERTM construct (4-OHT; diluted from 1 mM stock) (Sigma-Aldrich, catalog number: H7904 )
- SuperScript Vilo cDNA Synthesis Kit (Thermo Fisher Scientific, catalog number: 11754-050 )
Equipment
- 37 °C, 5% CO2 incubator (Thermo Fisher Scientific, Thermo ScientificTM, model: FormaTM Series II 3110 )
- Shaker (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 4625Q )
- Confocal laser scanning Microscope (Zeiss, models: LSM Meta 510 and 780 equipped with argon [488], helium-neon [543 and 633] and diode [405] lasers and Plan-Neofluar 40x DIC objective [NA = 1.3, oil])
- Zeiss Axio vert 200 microscope equipped with Apotome system (Zeiss) and 20x Plan apochromat objective (NA = 0.8, air), MRr digital camera and Axiovision 4.4 software.
- Light Cycler 480 II instrument (Roche Diagnostics, model: Light Cycler 480 II )
- Nanodrop (Thermo Fisher Scientific, Thermo Scientific, model: NanoDrop 8000 )
- Heraeus Multifuge 3SR Plus (DJB Labcare, catalog number: 75004371 ) with Swing Swing-out rotor 4 place (DJB Labcare, catalog number: 75006445 ) and Microtitre carrier for 4 microtitre plates (DJB Labcare, catalog number: 75006449 )
Software
- ImageJ software (National Institute of Health, version 1.50i)
Procedure
文章信息
版权信息
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Marques, E. and Klefström, J. (2016). Lentiviral shRNA Screen to Identify Epithelial Integrity Regulating Genes in MCF10A 3D Culture. Bio-protocol 6(23): e2050. DOI: 10.21769/BioProtoc.2050.
分类
癌症生物学 > 通用技术 > 细胞生物学试验 > 细胞转化
细胞生物学 > 基于细胞的分析方法 > 基因表达
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link