- EN - English
- CN - 中文
Preparation and Analysis of Crude Autolytic Enzyme Extracts from Staphylococcus aureus
制备并分析葡萄球菌属的天然自溶酶提取物
发布: 2015年12月20日第5卷第24期 DOI: 10.21769/BioProtoc.1687 浏览次数: 13836
评审: Arsalan DaudiAnonymous reviewer(s)
Abstract
The metabolism of the cell surface during bacterial cell division involves synthesis and degradation of peptidoglycan (PGN), the major component of the bacterial cell wall. Bacteria have to ensure that their surface remains capable of withstanding high turgor pressures and, simultaneously, that the PGN at their surface is concealed from receptors produced by the host innate immune system. For cell separation to occur, and for PGN to be kept concealed, “old” PGN is degraded by specific PGN hydrolases, also known as autolysins, that are found at the bacterial cell surface or that are secreted into the growth medium.
Bacterial PGN hydrolases are cell wall lytic enzymes that comprise a broad and diverse group of proteins. It is often difficult to assign a specific function to a PGN hydrolase mainly because an organism can have a large number of hydrolases with redundant activities and one hydrolase can have more than one enzymatic activity and participate in various cell processes (Vollmer et al. 2008). Bacillus subtilis has ca. 35 known or hypothetical PGN hydrolases, whereas Escherichia coli (E. coli) and Staphylococcus aureus (S. aureus) have, respectively, ca.16 and 19 PGN hydrolases (Vollmer, 2012; Heidrich et al., 2001; Singh et al., 2012).
PGN hydrolases can be classified in three main classes: glycosidases, amidases and peptidases. Glycosidases cleave the glycan backbone and are divided into N-acetylglucosaminidases and N-acetylmuramidases. Amidases cleave the linkage between the peptide chain and the N-acetylmuramic residue of the glycan chain. Peptidases, such as endopeptidases and carboxypeptidases, are able to cleave peptide bonds between different amino acids of the PGN stem peptide.
Here we describe a method to extract PGN hydrolases, which are non-covalently linked to the S. aureus cell wall (Vollmer, 2008). Analysis of extracts containing denatured PGN hydrolytic enzymes is performed by running a zymogram gel (a SDS-PAGE gel containing crude bacterial cell walls or substrate cells), which is then incubated in a non-denaturing buffer to allow renaturation of the PGN hydrolases. These renatured enzymes can then be identified through the production of clear bands that are observed where cell wall digestion has occurred. The protocol is divided into three steps: A) Preparation of the crude autolytic extracts from S. aureus cells; B) Preparation of substrate cells for gel zymograms; C) Analysis of crude autolytic extracts by gel zymography.
We also show that this method can be used to determine the absence or altered activity of PGN hydrolases produced by different S. aureus mutant strains.
Materials and Reagents
- Petri dishes (Sarstedt AG, catalog number: 82.1473 )
- 1 μl loops (Sarstedt AG, catalog number: 86.1567.010 )
- 25 ml glass (or plastic disposable) pipettes (Normax, catalog number: 4.5434334 )
- 50 ml Falcon tubes (Sarstedt AG, catalog number: 62.548.004 )
- 2 ml micro tubes (Sarstedt AG, catalog number: 72.691 )
- JA-14 centrifuge tubes (Thermo Fisher Scientific, NalgeneTM, catalog number: 3120-0250 )
- JA-20 centrifuge tubes (Thermo Fisher Scientific, NalgeneTM, catalog number: 3114-0050 )
- Spacer plates with 0.75 mm integrated spacers (Bio-Rad Laboratories, catalog number: 165-3310 )
- Mini-PROTEAN® Comb, 10-well, 0.75 mm (Bio-Rad Laboratories, catalog number: 165-3354 )
- Staphylococcus aureus strains to analyze regarding the cell wall lytic activity of their PGN hydrolases.
Note: In order to produce substrate cells, researchers may use the Staphylococcus aureus NCTC8325-4 strain, which has been cured from prophages and it can be obtained from BEI Resources under the reference NR-45937, and Micrococcus luteus DSM20030 strain, which can be obtained from DSMZ stock center. - Tryptic Soy Agar plates (TSA) (BD, Difco, catalog number: 236950 )
- Tryptic Soy Broth (TSB) (BD, Bacto, catalog number: 211825 )
- Luria Agar (Miller's LB agar) (LA) (Conda, catalog number: 1552 )
- Luria Broth (Miller's LB broth) (LB) (Conda, catalog number: 1551 )
- Ethanol (Merck Millipore Corporation, catalog number: 1.02371.1000 )
- Ice (homemade)
- Liquid nitrogen (Air Liquide)
- Tris (Trizma® base) (Sigma-Aldrich, catalog number: T1503 )
- Sodium chloride (Merck Millipore Corporation, catalog number: 1.06444.1000 )
- Hydrochloric acid (Merck Millipore Corporation, catalog number: 1.01834.2500 )
- Sodium Dodecyl Sulfate (Sigma-Aldrich, catalog number: L5750 )
- Glycine (Sigma-Aldrich, catalog number: G8898 )
- 30% Acrylamide/Bis Solution (Bio-Rad Laboratories, catalog number: 161-0158 )
- Dithiothreitol (DTT) (VWR International, catalog number: V3155 )
- Ammonium persulfate (APS) [(NH4)2S2O8] (Sigma-Aldrich, catalog number: A3678 )
- Bromophenol Blue sodium salt (Sigma-Aldrich, catalog number: B8026 )
- Precision Plus ProteinTM Dual Color Standards (PPPS) (Bio-Rad Laboratories, catalog number: 1610374 )
- Methylene Blue hydrate (Sigma-Aldrich, catalog number: 66720 )
- Triton X-100 (Sigma-Aldrich, catalog number: T8787 )
- Magnesium chloride hexahydrate (Sigma-Aldrich, catalog number: M9272 )
- Calcium chloride dihydrate (CaCl2.2H2O) (Sigma-Aldrich, catalog number: C3306 )
- Potassium hydroxide (KOH) (Sigma-Aldrich ,catalog number: P5958 )
- N, N, N′, N′-Tetramethylethylenediamine (TEMED) (Sigma-Aldrich, catalog number: T9281 )
- 70% ethanol (see Recipes)
- 500 mM Tris-HCl (pH 7.5)
(see Recipes)
- Washing buffer (see Recipes)
- 4% SDS (w/v) (see Recipes)
- 10% SDS (w/v) (see Recipes)
- 1.5 M Tris-HCl (pH 8.8)
(see Recipes)
- 0.5 M Tris-HCl (pH 6.8)
(see Recipes)
- 10% APS (w/v)
(see Recipes)
- Zymogram SDS-PAGE gels (see Recipes)
- 5x Laemmli loading buffer (see Recipes)
- Tris-Glycine-SDS buffer (see Recipes)
- Renaturation buffer (see Recipes)
- Methylene Blue solution (see Recipes)
Equipment
- Erlenmeyer flasks of 100 ml capacity (Normax, catalog number: 2121624N )
- Erlenmeyer flasks of 1 L capacity (Normax, catalog number: 2121654N )
- Cuvettes (Sarstedt AG, catalog number: 67.742 )
- 1.5 ml micro tubes (Sarstedt AG, catalog number: 72.690.001 )
- Micropipette (Gilson, model: Pipetman P1000 )
- Magnetic stir bars (VWR International, catalog number: 442-0361 )
- 30 °C/37 °C Incubator shaker (Eppendorf, New Brunswick Scientific, model: Innova® 40 )
- 30 °C /37 °C Incubator (BINDER GmbH, model: WTB )
- Burner
Note: In this work we used a wall-attached burner, but a portable burner also works. - Spectrophotometer (GE Healthcare, Amersham, model: Novaspec Plus Visible Spectrophotometer )
- Centrifuge (Beckman Coulter, model: Avanti J-26 XPI )
- JA-14 rotor (Beckman Coulter, model: 339247 )
- JA-20 rotor (Beckman Coulter, model: 334831 )
- Bench centrifuge (Eppendorf, model: 5430R )
- Rotor for 1.5 ml tubes (Thermo Fisher Scientific, Eppendorf, model: FA-45-24-11-HS )
- Shaker for 1.5 ml tubes (Eppendorf, model: Thermomixer® Comfort )
- Nanodrop (Thermo Fisher Scientific, model: ND-2000C )
- Autoclave
- Speed vac (Labconco, model: 78100 )
- Short plates (Bio-Rad Laboratories, catalog number: 165-3308 )
- Power Pac HV Power Supply (Bio-Rad Laboratories, catalog number: 164-5056 )
- Mini-PROTEAN Casting Stand Gaskets (Bio-Rad Laboratories, catalog number: 165-3305 )
- Mini-PROTEAN® Casting Frame (Bio-Rad Laboratories, catalog number: 165-3304 )
- Mini-PROTEAN® Casting Stand (Bio-Rad Laboratories, catalog number: 165-3303 )
- Mini-PROTEAN® Tetra Cell (Bio-Rad Laboratories, catalog number: 165-8000 )
Procedure
- Preparation of the crude autolytic extracts from S. aureus cells
- The working area was decontaminated with 70% ethanol (v/v) using paper towels and the burner was turned on. This procedure was done throughout the protocol every time sterile conditions were needed (Note 1).
- The bacterial S. aureus strains of interest were inoculated on TSA plates by streak plating using 1 μl loops and incubated overnight in a 30 °C incubator (Note 2).
- Under sterile conditions, a single isolated colony was picked with a 1 μl loop and inoculated in 10 ml of TSB in a 100 ml Erlenmeyer flask. Cultures were grown overnight (16 h-18 h) in a 30 °C incubator shaker at 200 rpm.
- The optical densities at λ = 600 nm of the overnight cultures were measured after making a 1/10 dilution (100 μl of culture in 900 μl of TSB) in a cuvette, under sterile conditions.
- The volume required for a starting OD600 of ~ 0.05 was taken from the overnight cultures and inoculated in 250 ml of TSB in a 1 L Erlenmeyer flask. The cultures were grown in a 30 °C incubator shaker at 200 rpm.
- The centrifuge JA-26 XPI was prepared by placing the JA-14 rotor and cooling it down to 4 °C.
- An ice/ethanol bath was prepared and kept at 4 °C: The ice was poured into a container and absolute ethanol was homogenously spread onto it. The flask of Washing buffer solution, the JA-14 and JA-20 centrifuge tubes were placed in it to cool down.
- When the cultures reached an OD600 ~ 0.3 (which should take about 3 h), they were put in the ice/ethanol bath (Note 3).
- The cultures were transferred into cold JA-14 centrifuge tubes. The tubes were balanced and the cells were pelleted at 4 ºC, 15,050 x g for 15 min.
- After centrifugation, the JA-14 rotor was replaced by the JA-20, which was cooled down to 4 °C.
- The supernatants were discarded by inversion of the tubes.
- The cells were resuspended in 20 ml of ice-cold Washing buffer and transferred to cold JA-20 centrifuge tubes. Washing buffer was added to the maximum capacity of the tubes (~46 ml).
- Cells were centrifuged as before (step A9).
- The supernatants were carefully removed using a 25 ml glass (or plastic) pipette until a semidry pellet was left at the bottom of the tubes.
- The cells were carefully resuspended in 250 μl of 4% SDS (w/v), transferred to a 1.5 ml micro tube and incubated in a thermomixer at 25 °C, 700 rpm for 30 min (Note 4).
- After incubation, the cells were harvested using a bench centrifuge at 22,380 x g, for 15 min, room temperature (RT).
- The supernatants, containing the crude enzyme autolytic extracts, were transferred to clean micro tubes.
- The extracts were quantified in the Nanodrop (Proteins-280 nm) using a 2 μl aliquot. Mili-Q water was used as a blank.
- Each extract was divided into 50 μl aliquots, frozen in liquid nitrogen and stored at -80 °C until further use (Note 5).
- The working area was decontaminated with 70% ethanol (v/v) using paper towels and the burner was turned on. This procedure was done throughout the protocol every time sterile conditions were needed (Note 1).
- Preparation of substrate cells for gel zymograms
- For preparation of substrate cells, the S. aureus NCTC8325-4 and the M. luteus DSM20030 strains were plated on TSA and LA plates, respectively, by streak plating using 1 μl loops, and incubated in a 37 °C incubator. S. aureus was incubated overnight and M. luteus for at least 24 h (Note 2).
- Under sterile conditions, a single isolated colony of each strain was picked with a 1 μl loop and inoculated into 10 ml of TSB or LB in 100 ml Erlenmeyer flasks. Cultures were grown overnight (16 h-18 h) in a 37 °C incubator shaker at 200 rpm.
- The optical densities at λ = 600 nm of the overnight cultures were measured after 1/10 dilution, using as blanks 1 ml of the sterile TSB or LB.
- The volume required for a starting OD600 of ~ 0.05 was taken from the overnight cultures and inoculated in 250 ml of TSB or LB in a 1 L Erlenmeyer flask. The cultures were grown using a 37 °C incubator shaker at 200 rpm.
- When the cultures reached an OD600 ~ 1.0 the cells were harvested by centrifugation in a JA-14 tube for 15 min at 15,050 x g at room temperature. M. luteus culture should take one full day while S. aureus should take about 3 h to reach the desired OD (Note 6).
- The supernatants were discarded and cells were washed with 250 ml of Milli-Q water.
- The cells were centrifuged as above and the supernatants were discarded.
- The cells were resuspended in 30 ml of Milli-Q water and transferred to 50 ml Falcon tubes.
- The cell suspension was autoclaved for 15 min, 121 °C (Note 7).
- The autoclaved cells were transferred to JA-20 tubes and centrifuged for 15 min at 15,050 x g, RT.
- The supernatants were discarded and pellets were stored overnight at -20 °C (Note 8).
- The pellets were defrosted at room temperature, resuspended in 3 ml of Milli-Q water and divided in half into two pre-weighted 2 ml tubes.
- The suspension was lyophilized overnight in the speed vac.
- The weight of the tubes containing the cells was determined and the dry weight was calculated.
- Bacterial substrate cells were resuspended thoroughly in Milli-Q water at 50 mg/ml final concentration and stored at -20 °C (Note 9).
- For preparation of substrate cells, the S. aureus NCTC8325-4 and the M. luteus DSM20030 strains were plated on TSA and LA plates, respectively, by streak plating using 1 μl loops, and incubated in a 37 °C incubator. S. aureus was incubated overnight and M. luteus for at least 24 h (Note 2).
- Analysis of the crude autolytic extracts by gel zymography
- Substrate cells were thawed and, to ensure a complete resuspension, kept overnight at room temperature with agitation using a magnetic stirrer at maximum speed (Note 10).
- Two resolving Acrylamide/0.2% Bisacrylamide SDS-PAGE gels (Laemmli, 1970) were prepared containing a final concentration of 2 mg/ml of substrate cells from S. aureus (10 % gel) and M. luteus (8% gel) (Note 11 and Figure 1).
- A standard stacking gel, lacking substrate cells, was placed on top of the resolving gels (Laemmli, 1970).
- 10 μg (for the M. luteus gel) and 15 μg (for the S. aureus gel) of crude autolytic enzyme extracts (Note 12) were mixed with Laemmli loading buffer (Laemmli, 1970) at a final concentration of 2x (samples were not heated, see Recipes).
- The electrophoresis conditions used were as follows:
- Temperature: Room Temperature.
- Voltage constant: 70 V (per gel).
- Running buffer: Tris-Glycine-SDS buffer.
- Temperature: Room Temperature.
- After electrophoresis, the gels were rinsed once with Milli-Q water.
- The gels were washed 3 times with Milli-Q for 15 min at RT with gentle agitation.
- The gels were incubated overnight at 37 °C with gentle agitation in Renaturation buffer (Note 13).
- Zymograms were stained in Methylene Blue Solution for an hour and destained in water until the bands were clear (Note 14 and Figure 2).
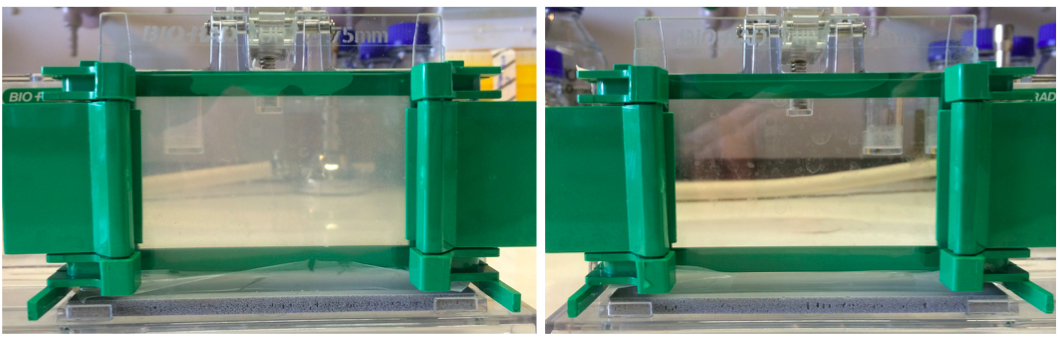
Figure 1. A 10% zymogram gel and SDS-PAGE gel. Left panel shows a zymogram gel, which is slightly opaque. Right panel shows a regular SDS-PAGE gel that is completely transparent due to the lack of substrate cells.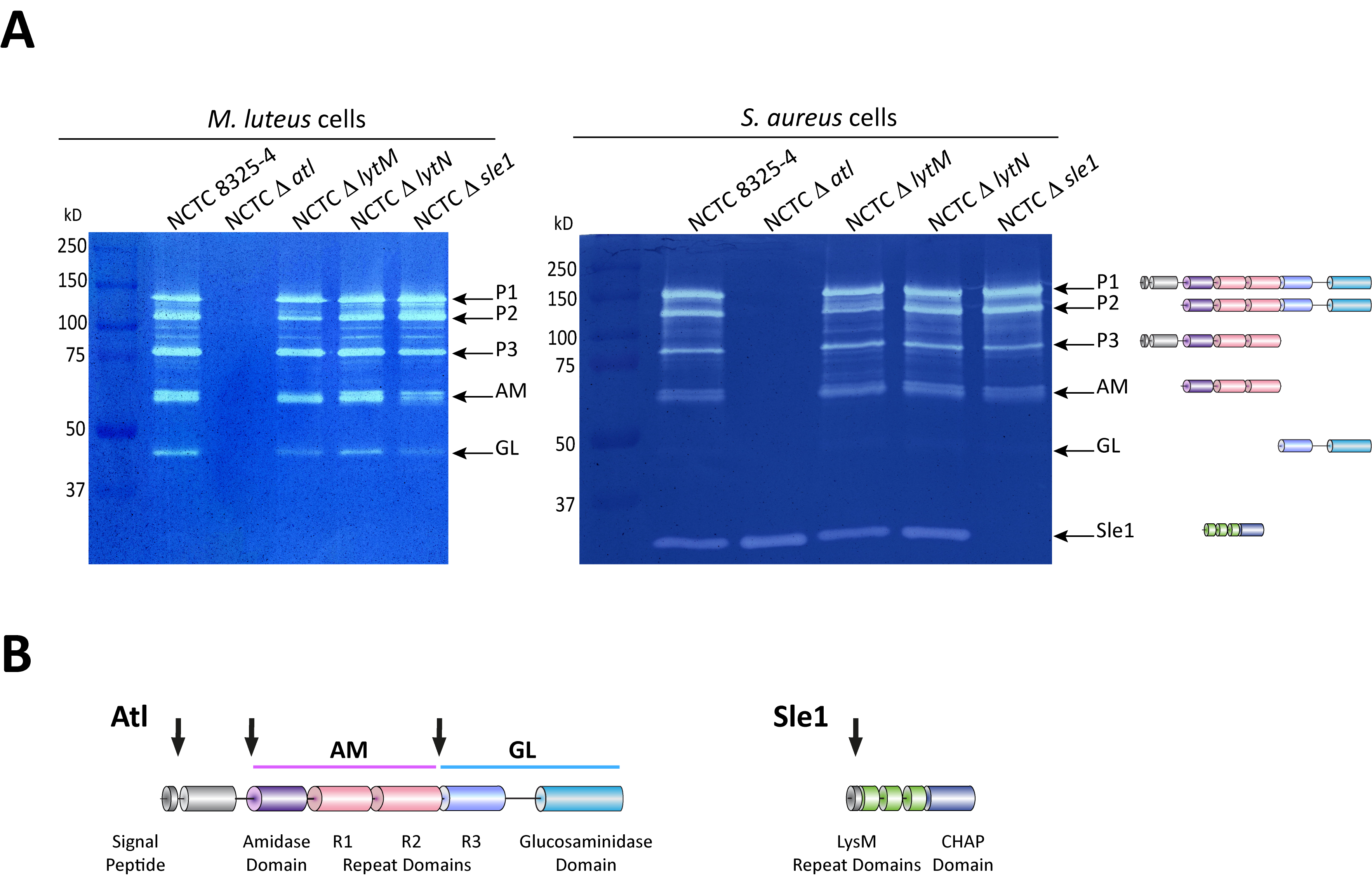
Figure 2. Zymography analysis of S. aureus autolytic enzymes. A. Gel zymography analysis of crude autolytic enzyme extracts of S. aureus. M. luteus and S. aureus zymogram gels were used to analyze the autolytic enzymatic extracts harvested from the parental S. aureus NCTC8325-4 strain and from mutants that lack different autolysins (NCTCΔatl, which lacks the major Atl autolysin; NCTCΔlytM, which lacks the glycyl-glycine endopeptidase LytM; NCTCΔlytN, which lacks the putative cell wall hydrolase LytN and NCTCΔsle1, which lacks the N-acetylmuramoyl-L-alanine amidase Sle1). Five different forms of the major autolysin Atl were seen with either substrate cells. However, only the S. aureus substrate cells allowed detection of Sle1 activity, hence showing Sle1 substrate specificity for S. aureus peptidoglycan. B. Representation of the post-translational cleavage (black arrows) of the atl encoded protein (left). The processed amidase (AM) releases stem peptides from PGN by cutting the bond between the stem peptides and the N-acetylmuramic acid residue. The glucosaminidase (GL) releases muropeptides by cutting the glycosidic linkage between N-acetylmuramic acid and N-acetylglucosamine of glycan strands. Representation of Sle1 (right), which includes three LysM domains (responsible for the interaction with PGN) and a CHAP domain (associated with amidase activity of different PGN hydrolases).
- Substrate cells were thawed and, to ensure a complete resuspension, kept overnight at room temperature with agitation using a magnetic stirrer at maximum speed (Note 10).
Notes
- The working area should be a safe place to work near a flame, i.e., no air currents and away from explosives, flammable materials and solvents. For this protocol, work next to the flame can be substituted by working in a biological safety cabinet. Diligently follow all recommended safety guidelines and waste disposal regulations recommended by your host institution.
- Plating of the strains should be done from a glycerol stock onto a fresh media plate to ensure the viability of bacterial culture and to prevent selection of suppressor mutants. Growth of the S. aureus strains used in this work is not affected by light/dark conditions.
- Incubating the bacterial culture to an OD600 ~ 0.3 ensures that bacteria are harvested during the exponential growth phase. The fast transfer of bacteria to the ice/ethanol bath will reduce the activity of autolysins, which would modify the PGN present at the surface of the substrate cells, and prevent lysis of bacteria, which would result in the loss of specific PGN hydrolases to the growth medium.
- This step is required to extract proteins that are associated with the bacterial cell surface without inducing the lysis of bacteria.
- Dividing the extract in different aliquots will prevent the consecutive thawing and freezing steps that would result in the loss of activity of certain PGN hydrolases (3 cycles of thawing and freezing did not result in loss of cell wall lytic activity of Atl and Sle1).
- To harvest S. aureus and M. luteus at the same time please take in account their different growth rates.
- This step ensures that enzymes in this cell suspension lose their activity, and therefore do not cause lysis of the substrate cells during gel zymography analysis.
- We have obtained better results when autoclaved cells were stored at -20 °C prior the step of lyophilization. We assume that freezing step may help the resuspension of the substrate.
- The use of different bacteria as substrate cells in the gel zymography analysis may result in differences in the intensity and number of bands observed in the zymogram.
- This is a critical step as if substrate cells are not well resuspended, the resulting zymogram will be grainy, lowering the sensitivity of the gel zymography analysis.
- The M. luteus cell wall is considerably more sensitive to PGN lytic enzymes than S. aureus cells. Therefore when M. luteus cells are used, there is an increase in the intensity and in the number of bands that are detected in a Zymogram. It is preferable to use an 8% gel when using in M. luteus as substrate cells so that there is a better separation of the high molecular weight. In S. aureus gels, it is preferable to run a more concentrated gel to detect the activity of Sle1 that runs close to the 37 kDa protein standard.
- Also due to the high susceptibility of M. luteus cells to PGN hydrolases, the optimum quantity of extracts protein is 5-10 μg whereas for S. aureus gel the minimum extracts protein amount to be used should be 15 μg.
- This step is required to regain the activity of the PGN hydrolases, which was lost during treatment with the SDS solution.
- In this step, clear bands are observed due to the presence of autolysins. Intact substrate cells will retain the dye, while no color will be observed where substrate bacterial cells have lysed.
In the gel containing M. luteus cells, the clear bands start to be seen after a 15 min wash and a picture of the gel should be taken before changing the water. The washing step should be repeated twice, taking pictures of the gel with every change of the water.
The gel with S. aureus cells will take at least three 15 min washes before the bands start to be seen. We usually take a picture after three washes, then after a fourth or fifth wash of about 3 h-5 h. A final overnight wash is recommended to destain the gel as much as possible so that less active lytic proteins may be better seen (e.g. GL activity).
Recipes
- 70% ethanol
Into a spray bottle, mix 70 ml of ethanol with 30 ml of water - 500 mM Tris-HCl (pH 7.5)
Add 30.3 g of Trizma® base to 300 ml of Milli-Q water
Let it stir until solubilisation occurs
Adjust the pH to 7.5 with 1 M HCl
Fill up to 500 ml with Mili-Q water
Confirm pH and autoclave
Use it as a sterile stock solution
Stored at RT - Washing buffer
50 mM Tris-HCl (pH 7.5)/150 mM NaCl
To 4.4 g of NaCl add 50 ml of 500 mM Tris-HCl (pH 7.5) and 400 ml of Milli-Q water
After solubilisation, confirm pH and add Milli-Q water to 500 ml
Filter sterilize (0.2 μm) or autoclave
Stored at RT until use - 4% SDS (w/v)
Dissolve 4 g of SDS in 80 ml of Milli-Q water by stirring
SDS is an irritant agent
Wear a mask while weighting the powder
Fill up to 100 ml with Mili-Q water
Filter sterilize (0.2 μm)
Stored at RT - 10% SDS (w/v)
Weight 10 g and dissolve in 80 ml of Milli-Q water by stirring
SDS is an irritant agent
Wear a mask while weighting the powder
Fill up to 100 ml with Milli-Q water
Filter sterilize (0.2 μm)
Stored at RT - 1.5 M Tris-HCl (pH 8.8)
Add 18.2 g of Trizma® base to 60 ml of Milli-Q water
Let it stir until solubilisation occurs
Adjust the pH to 8.8 with 1 M HCl
Fill up to 100 ml with Milli-Q water
Confirm pH and filter sterilize (0.2 um)
Keep it as a sterile stock solution-use an aliquot (~20 ml) as a working solution
Stored at RT - 0.5 M Tris-HCl (pH 6.8)
Add 6.06 g of Trizma® base to 60 ml of Milli-Q water
Let it stir until solubilisation occurs
Adjust the pH to 6.8 with 1 M HCl
Fill up to 100 ml with Mili-Q water
Confirm pH and filter sterilize (0.2 um)
Keep it as a sterile stock solution-use an aliquot (~20 ml) as a working solution.
Stored at RT - 10% APS (w/v)
Weight 1 g and dissolve in 10 ml of Milli-Q water
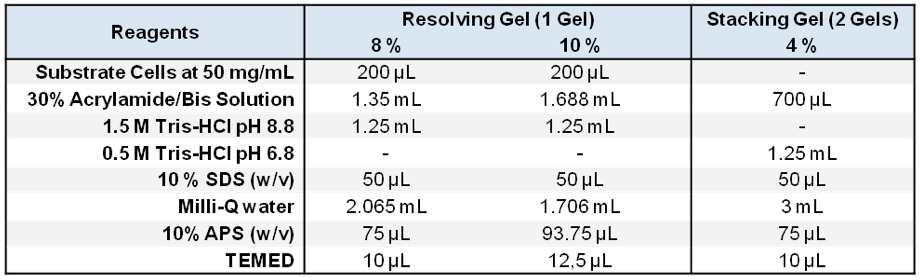
Stored in 500 μl aliquots at -20 °C - Zymogram SDS-PAGE gels
- 5x Laemmli loading buffer
Weight 0.77 g DTT, 4.3 g Glycerol, 2 mg Bromophenol Blue
Add 5 ml 0.5 M Tris (pH 6.8) and fill up to 10 ml with Milli-Q water
Stored at -20 °C in 50 μl aliquots - Tris-Glycine-SDS buffer
Dissolve 30.0 g of Tris base, 144.0 g of glycine, and 10.0 g of SDS in 1,000 ml of water
Stored at RT - Renaturation buffer
50 mM Tris-HCl (pH 7.5), 0.1% (v/v) Triton X-100, 10 mM CaCl2, 10 mM MgCl2:
Weight 0.74 g of calcium chloride dihydrate and 1.02 g of magnesium chloride hexahydrate
Take 50 ml of 500 mM Tris-HCl (pH 7.5) and 500 μl of Triton X-100
Stir until solubilisation
Always use fresh - Methylene blue solution
0.1% (w/v) methylene blue in 0.01% potassium hydroxide:
Dissolve 0.5 g of methylene blue and 0.05 g of potassium hydroxide in 500 ml Milli-Q water
Stored at RT
Acknowledgments
This protocol, which was adapted or modified from Grilo et al. (2014) and Yokoi et al. (2008), has been used to describe that autolysins can prevent detection of the PGN at the bacterial cell surface (Atilano et al., 2014). This work was supported by fellowship SFRH/BD/78748/2011 to FV and project PTDC/BIA-PLA/3432/2012 to SRF from Fundação para a Ciência e Tecnologia. We thank Inês Grilo for suggestions and Teresa Baptista da Silva for technical support.
References
- Atilano, M. L., Pereira, P. M., Vaz, F., Catalao, M. J., Reed, P., Grilo, I. R., Sobral, R. G., Ligoxygakis, P., Pinho, M. G. and Filipe, S. R. (2014). Bacterial autolysins trim cell surface peptidoglycan to prevent detection by the Drosophila innate immune system. Elife 3: e02277.
- Gotz, F., Heilmann, C. and Stehle, T. (2014). Functional and structural analysis of the major amidase (Atl) in Staphylococcus. Int J Med Microbiol 304(2): 156-163.
- Grilo, I. R., Ludovice, A. M., Tomasz, A., de Lencastre, H. and Sobral, R. G. (2014). The glucosaminidase domain of Atl - the major Staphylococcus aureus autolysin - has DNA-binding activity. Microbiologyopen 3(2): 247-256.
- Heidrich, C., Templin, M. F., Ursinus, A., Merdanovic, M., Berger, J., Schwarz, H., de Pedro, M. A. and Holtje, J. V. (2001). Involvement of N-acetylmuramyl-L-alanine amidases in cell separation and antibiotic-induced autolysis of Escherichia coli. Mol Microbiol 41(1): 167-178.
- Laemmli, U. K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227(5259): 680-685.
- Oshida, T., Sugai, M., Komatsuzawa, H., Hong, Y. M., Suginaka, H. and Tomasz, A. (1995). A Staphylococcus aureus autolysin that has an N-acetylmuramoyl-L-alanine amidase domain and an endo-beta-N-acetylglucosaminidase domain: cloning, sequence analysis, and characterization. Proc Natl Acad Sci U S A 92(1): 285-289.
- Singh, S. K., SaiSree, L., Amrutha, R. N. and Reddy, M. (2012). Three redundant murein endopeptidases catalyse an essential cleavage step in peptidoglycan synthesis of Escherichia coli K12. Mol Microbiol 86(5): 1036-1051.
- Yokoi, K. J., Sugahara, K., Iguchi, A., Nishitani, G., Ikeda, M., Shimada, T., Inagaki, N., Yamakawa, A., Taketo, A. and Kodaira, K. (2008). Molecular properties of the putative autolysin Atl(WM) encoded by Staphylococcus warneri M: mutational and biochemical analyses of the amidase and glucosaminidase domains. Gene 416(1-2): 66-76.
- Vollmer, W., Joris, B., Charlier, P. and Foster, S. (2008). Bacterial peptidoglycan (murein) hydrolases. FEMS Microbiol Rev 32(2): 259-286.
- Vollmer, W. (2012). Bacterial growth does require peptidoglycan hydrolases. Mol Microbiol 86(5): 1031-1035.
文章信息
版权信息
Vaz and Filipe. This article is distributed under the terms of the Creative Commons Attribution License (CC BY 4.0).
如何引用
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Vaz, F. and Filipe, S. R. (2015). Preparation and Analysis of Crude Autolytic Enzyme Extracts from Staphylococcus aureus. Bio-protocol 5(24): e1687. DOI: 10.21769/BioProtoc.1687.
- Atilano, M. L., Pereira, P. M., Vaz, F., Catalao, M. J., Reed, P., Grilo, I. R., Sobral, R. G., Ligoxygakis, P., Pinho, M. G. and Filipe, S. R. (2014). Bacterial autolysins trim cell surface peptidoglycan to prevent detection by the Drosophila innate immune system. Elife 3: e02277.
分类
微生物学 > 微生物生物化学 > 蛋白质 > 活性
微生物学 > 微生物细胞生物学 > 基于细胞的分析方法
生物化学 > 糖类 > 糖蛋白
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link