往期刊物2014
卷册: 4, 期号: 7
生物化学
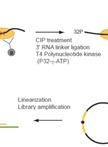
Individual-nucleotide-resolution UV Cross-linking and Immunoprecipitation (iCLIP) of UPF1
UPF1蛋白单核苷酸分离紫外线交联和免疫沉淀(iCLIP)
UPF1 RNA Immunoprecipitation from Mini-μ Construct–expressing Cells
Mini-μ 载体-表达细胞中的UPF1 RNA 免疫沉淀
癌症生物学
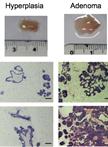
Isolation and Immortalization of Fibroblasts from Different Tumoral Stages
不同肿瘤阶段成纤维细胞的分离和永生化
免疫学
Isolation of Cells from Human Intestinal Tissue
从人小肠组织分离细胞
α2β1-integrin Clustering and Internalization Protocol
α2β1-整合素的群集和内化实验方案
微生物学
Flow Cytometric Analysis of Autophagic Activity with Cyto-ID Staining in Primary Cells
使用Cyto-ID染色原代细胞并用流式细胞法分析其自噬活性
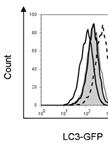
Flow Cytometric Analyses of Autophagic Activity using LC3-GFP fluorescence
采用LC3B-GFP荧光对自噬活性进行流式细胞检测分析
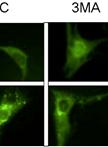
Fluorescence Microscopy Analysis of Drug Effect on Autophagosome Formation
荧光显微法分析药物对自噬体形成的影响
Native BAD-1 Binding to Heparin-agarose
天然BAD-1与肝素琼脂糖的结合
分子生物学
Electrophoresis Mobility Shift Assay
电泳迁移率实验
植物科学
Quantification of Anthocyanin Content
花青素含量的定量测定
Seed Coat Ruthenium Red Staining Assay
种皮钌红染色试验
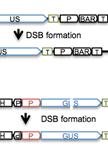
Measuring Homologous Recombination Frequency in Arabidopsis Seedlings
拟南芥幼苗中同源重组频率的测定
DNA Damage Sensitivity Assays with Arabidopsis Seedlings
拟南芥幼苗的DNA损伤敏感性实验
干细胞
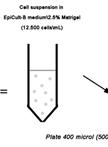
3D Mammary Colony-Forming Cell Assay
3D 乳腺集落形成细胞培养方法