往期刊物2016
卷册: 6, 期号: 3
癌症生物学
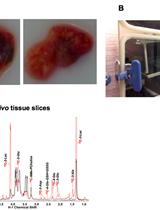
Stable Isotope Resolved Metabolomics Studies in ex vivo TIssue Slices
体外组织切片的稳定同位素分解代谢组学研究
细胞生物学
Mouse Oocyte Isolation, Cultivation and RNA Microinjection
小鼠卵母细胞分离、培养和RNA显微注射
免疫学
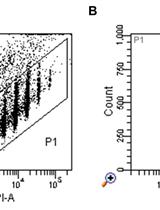
Mouse BMDC-dependent T Cell Polarization Assays
小鼠BMDC依赖性T淋巴细胞的极化试验
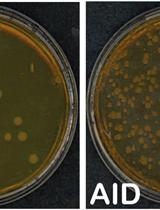
Cell-based Assays to Monitor AID Activity
基于细胞的监测AID活性的方法
微生物学
Design and Functional Analysis of Fluorescent Nitrate and Peptide Transporter Activity Sensors in Yeast Cultures
酵母培养物中荧光硝酸盐和肽转运载体传感器的设计和功能分析
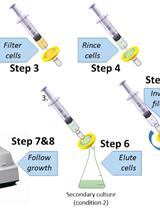
Calculation of Microorganism Lag Times as a Measure of Adaptative Capability between Different Growth Conditions
采用微生物滞后时间计算衡量不同生长条件下的自适应能力
神经科学
Craniotomy for Cortical Voltage-sensitive Dye Imaging in Mice
开颅手术后利用电压敏感性染料进行小鼠脑皮层成像
植物科学
Structured Illumination Microscopy (SIM) and Photoactivated Localization Microscopy (PALM) to Analyze the Abundance and Distribution of RNA Polymerase II Molecules on Flow-sorted Arabidopsis Nuclei
使用结构照明显微镜(SIM)和光敏定位显微镜(PALM)分析RNA聚合酶II分子在流式分选的拟南芥细胞核中的丰度及分布
Quantification of Ethylene Production in Tomato Leaves Infected by Xanthomonas euvesicatoria
黄单胞菌感染番茄叶片后乙烯产生的定量测定
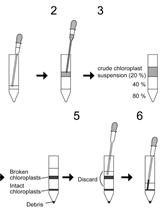
Preparation of Chloroplast Lipid Membrane and Lipid-protein Interaction Assay
叶绿体膜脂的制备及脂质-蛋白质相互作用的分析
Measurement of PI4P Levels in Intact Chloroplasts Isolated from Arabidopsis thaliana
测量从拟南芥中分离的完整叶绿体中的PI4P含量
干细胞
Transfection of Embryoid Bodies with miRNA Precursors to Induce Cardiac Differentiation
用miRNA前体转染拟胚体来诱导心肌分化