往期刊物2022
卷册: 12, 期号: 6
生物化学
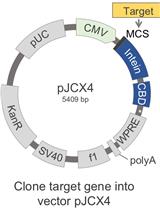
Novel Protein Production Method Combining Native Expression in Human Cells with an Intein-based Affinity Purification and Self-cleavable Tag
将人类细胞中的天然表达与基于内含肽的亲和纯化和自切割标签相结合的新型蛋白质生产方法
Analysis of Heterocyst and Akinete Specific Glycolipids in Cyanobacteria Using Thin-layer Chromatography
使用薄层色谱法分析蓝藻中的异囊体和动体特异性糖脂
生物工程
A Fluorescence-based Assay for Measuring Phospholipid Scramblase Activity in Giant Unilamellar Vesicles
荧光法测定巨大单层囊泡中磷脂爬行酶活性的方法
生物物理学
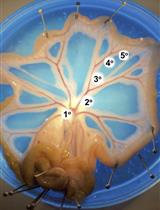
Electrophysiological Measurements of Isolated Blood Vessels
离体血管的电生理测量
癌症生物学
Single-cell Damagenome Profiling by Linear Copying and Splitting based Whole Genome Amplification (LCS-WGA)
基于线性复制和分裂的全基因组扩增法(LCS-WGA)分析单细胞损伤组
Small Molecule Screening of Primary Human Acute Myeloid Leukemia Using Co-culture and Multiplexed FACS Analysis
使用共培养和多重 FACS 分析对原发性人类急性髓细胞白血病进行小分子筛选
发育生物学
3D-Structured Illumination Microscopy of Centrosomes in Human Cell Lines
人类细胞系中心体的 3D 结构照明显微镜
TUNEL Labeling to Detect Double-stranded DNA Breaks in Caenorhabditis elegans Gonads
TUNEL 标记检测秀丽隐杆线虫性腺中的双链 DNA 断裂
Measurement of Contractile Ring Tension Using Two-photon Laser Ablation during Drosophila Cellularization
果蝇细胞化过程中使用双光子激光消融测量收缩环张力
免疫学
Differentiation of Human Induced Pluripotent Stem Cell into Macrophages
人诱导多能干细胞向巨噬细胞的分化
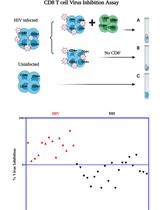
CD8 T Cell Virus Inhibition Assay Protocol
CD8 T 细胞病毒抑制实验方法
微生物学
Generation, Maintenance and HBV Infection of Human Liver Organoids
人肝类器官的产生、维持和HBV感染
Tracking the Reversed Oxidative Tricarboxylic Acid Cycle in Bacteria
追踪细菌中的反向氧化三羧酸循环
Three-dimensional Models of the Nasopharynx for the Study of Epstein-Barr Virus Infection
研究EB病毒感染的鼻咽三维模型
Phytophthora sojae Transformation Based on the CRISPR/Cas9 System
基于CRISPR/Cas9系统的大豆疫霉转化
植物科学
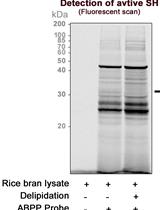
Activity-based Protein Profiling of Serine Hydrolase Superfamily Enzymes
丝氨酸水解酶超家族酶的基于活性的蛋白质分析