往期刊物2021
卷册: 11, 期号: 9
生物化学
Click Chemistry for Imaging in-situ Protein Palmitoylation during the Asexual Stages of Plasmodium falciparum
恶性疟原虫无性期原位蛋白质棕榈酰化的点击化学成像
Chromatographic Assays for the Enzymatic Degradation of Chitin
几丁质酶解的色谱分析
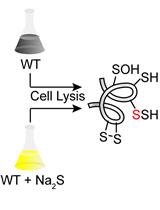
Proteomics Profiling of S-sulfurated Proteins in Acinetobacter baumannii
鲍曼不动杆菌硫酸化蛋白的蛋白质组学分析
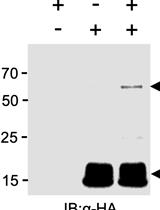
Characterising Plant Deubiquitinases with in vitro Activity-based Labelling and Ubiquitin Chain Disassembly Assays
植物去泛素酶的体外活性标记和泛素链分解分析
生物工程
Inkjet 3D Printing of Polymers Resistant to Fungal Attachment
抗真菌附着聚合物的喷墨3D打印
生物物理学
Spin Labeling of RNA Using “Click” Chemistry for Coarse-grained Structure Determination via Pulsed Electron-electron Double Resonance Spectroscopy
利用脉冲电子-电子双共振光谱测定粗粒结构的“点击”化学的RNA自旋标记
癌症生物学
ATAC-Seq-based Identification of Extrachromosomal Circular DNA in Mammalian Cells and Its Validation Using Inverse PCR and FISH
基于ATAC-Seq的哺乳动物细胞染色体外环状DNA鉴定及其反向PCR和FISH验证
免疫学
Analysis of Monocyte Cell Fate by Adoptive Transfer in a Murine Model of TLR7-induced Systemic Inflammation
TLR7诱导的系统性炎症小鼠模型中通过过继转移单核细胞命运分析
微生物学
COVID-19 Sample Pooling: From RNA Extraction to Quantitative Real-time RT-PCR
COVID-19样本汇集:从RNA提取到实时定量RT-PCR
High-throughput Method for Detecting Siderophore Production by Rhizosphere Bacteria
根际细菌产生铁载体的高通量检测方法
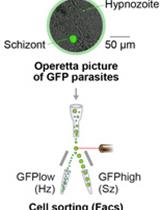
Isolation of GFP-expressing Malarial Hypnozoites by Flow Cytometry Cell Sorting
流式细胞术分离表达GFP的疟原虫催眠素
Assessing Swarming of Aerobic Bacteria from Human Fecal Matter
人类粪便中需氧细菌群集的评估
分子生物学
Identification of R-loop-forming Sequences in Drosophila melanogaster Embryos and Tissue Culture Cells Using DRIP-seq
DRIP-seq法鉴定黑腹果蝇胚胎和组织培养细胞R-loop形成序列
神经科学
Activity-based Anorexia for Modeling Vulnerability and Resilience in Mice
基于活动的厌食症小鼠脆弱性和恢复力模型的建立
植物科学
Histological Methods to Detect Early-stage Plant Defense Responses during Artificial Inoculation of Lolium perenne with Epichloë festucae
羊茅人工接种多年生黑麦草过程中植物早期防御反应的组织学检测
Age, Wound Size and Position of Injury – Dependent Vascular Regeneration Assay in Growing Leaves
年龄、伤口大小和损伤部位 – 生长叶片中依赖于损伤的维管再生试验