往期刊物2019
卷册: 9, 期号: 7
微生物学
Delivering "Chromatic Bacteria" Fluorescent Protein Tags to Proteobacteria Using Conjugation
利用接合生殖将产色细菌荧光蛋白标签导入变形杆菌
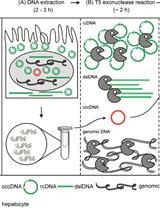
Quantification of Hepatitis B Virus Covalently Closed Circular DNA in Infected Cell Culture Models by Quantitative PCR
qPCR定量分析感染细胞培养模型中乙型肝炎病毒共价闭合环状DNA
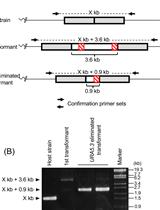
Multiple Modification of Chromosomal Loci Using URA5.3 Selection Marker in the Unicellular Red Alga Cyanidioschyzon merolae
在单细胞红藻 Cyanidioschyzon merolae中利用URA5.3选择标记对染色体基因进行多重修饰
分子生物学
Identification of RNase-sensitive LINE-1 Ribonucleoprotein Interactions by Differential Affinity Immobilization
利用差异化亲和固定鉴定核糖核酸酶敏感的LINE-1核糖核蛋白间的相互作用
神经科学
Consummatory Successive Negative Contrast in Rats
大鼠完成连续阴性对照实验
植物科学
Quantitative Plasmodesmata Permeability Assay for Pavement Cells of Arabidopsis Leaves
拟南芥叶片扁平细胞中胞间连丝渗透性定量测定实验
Visualization of Plant Cell Wall Epitopes Using Immunogold Labeling for Electron Microscopy
利用电子显微镜免疫金标记法进行植物细胞壁表位的可视化观察
Extraction and Purification of Laccases from Rice Stems
水稻茎秆酶的提取与纯化
Quantification of the Humidity Effect on HR by Ion Leakage Assay
离子渗漏实验定量分析湿度对超敏反应的影响