- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Chitinase Assay from Cultured Bone Marrow Derived Macrophages
Published: Vol 3, Iss 23, Dec 5, 2013 DOI: 10.21769/BioProtoc.983 Views: 8282
Reviewed by: Fanglian He

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Fluorescence Polarization-Based High-Throughput Screening Assay for Inhibitors Targeting Cathepsin L
Keyu Guo [...] Shuyi Si
Jul 20, 2025 2282 Views

Novel Experimental Approach to Investigate Immune Control of Vascular Function: Co-culture of Murine Aortas With T Lymphocytes or Macrophages
Taylor C. Kress [...] Eric J. Belin de Chantemèle
Sep 5, 2025 3550 Views

Detecting the Activation of Endogenous Small GTPases via Fluorescent Signals Utilizing a Split mNeonGreen: Small GTPase ActIvitY ANalyzing (SAIYAN) System
Miharu Maeda and Kota Saito
Jan 5, 2026 496 Views
Abstract
Chitinases are chitin-degrading enzymes. Chitinases play essential roles in combating chitin-containing pathogens as well as established roles in asthmatic inflammation. This assay is designed to detect chitinase activity in macrophage cell lysates. The chitin substrate is labeled with 4-methylumbelliferone. Hydrolysis of chitin releases 4-methylumbelliferone, and is measured fluorometrically to determine chitinase activity.
Materials and Reagents
- Cells to be analyzed. e.g. Bone Marrow-derived macrophages from C57BL/6 mice
- L-cell
- cDMEM/F12
- 4-Mehtylumbelliferyl labeled substrates:
- 4-Methylumbelliferyl N-acetyl-β-D-glucosaminide (exochitinase activity; β–N-acetylglucosaminidase activity) (Sigma-Aldrich, catalog number: M2133 )
- 4-Methylumbelliferyl N,N′-diacetyl-β-D-chitobioside (exochitinase activity; chitobiosidase activity) (Sigma-Aldrich, catalog number: M9763 )
- 4-Methylumbelliferyl β-D-N,N′,N′′-triacetylchitotriose (endochitinase activity) (Sigma-Aldrich, catalog number: M5639 )
- 4-Methylumbelliferyl N-acetyl-β-D-glucosaminide (exochitinase activity; β–N-acetylglucosaminidase activity) (Sigma-Aldrich, catalog number: M2133 )
- Chitinase from Trichoderma viride (Sigma-Aldrich, catalog number: C6242 )
- Methylumbelliferone Standard (50 mg/ml) (Sigma-Aldrich, catalog number: M1381 )
- PBS
- DMSO
- Plate reader
- Diabasic Sodium Phosphate
- Citric Acid
- Glycine
- DTT
- Assay buffer (see Recipes)
- Stop buffer (see Recipes)
- Protein lysis buffer (see Recipes)
- Substrate Stock (see Recipes)
- Positive control (see Recipes)
Equipment
- Flat bottom black 96-well plates
- Fluorescent plate reader (Molecular Devices)
Procedure
- Cells to be analyzed. We have used Bone Marrow-derived macrophages from C57BL/6 mice.
- In brief, bone marrow is flushed from tibias and femurs of 6-8 week old mice with 10 ml complete DMEM/F12 supplemented with 20% L-cell supernatant (day 0).
- Bone marrow cells are plated in non tissue culture treated 10 cm petri dishes ~10 ml cells per dish. After 4 days, add 10 ml additional media.
- Cells are harvested by gentle scraping on day 7.
- Macrophages are plated at 50,000 cells/well in 96 well plates in cDMEM/F12 in 10% L-cell supernatant and allowed to rest for 3 days.
- On day 10, media is changed to cDMEM/F12 without L-cell supernatant and rested overnight.
- Cells are ready for use on day 11.
- In brief, bone marrow is flushed from tibias and femurs of 6-8 week old mice with 10 ml complete DMEM/F12 supplemented with 20% L-cell supernatant (day 0).
- Aspirate media in each well from the culture plates, and add lysis buffer to each (50 μl for 96 well plate, and 200 μl for a 24 well plate). Place the plate on the rocker for 15 min at room temperature.
- Dilute an aliquot of 4-Methylumbelliferyl substrate stock solution 40-fold in assay buffer, such that the final concentration of the substrate is 0.5 mg/ml. This will be termed the “working solution”.
- Approximately 100 μl of working solution will be needed per sample. Allow solution to equilibrate in 37 °C water bath.
- For each form of chitinase being tested there is a unique substrate. Separate assay plates will needed for each substrate.
- Approximately 100 μl of working solution will be needed per sample. Allow solution to equilibrate in 37 °C water bath.
- Prepare Methylumbelliferone standards by diluting the top Methylumbelliferone standard 1:100 (500 μg/ml), 1:1,000 (50 μg/ml), and 1:10,000 (5 μg/ml) in stop buffer.
- For best resolution, add 2 μl top Methylumbelliferone standard to 198 μl of stop buffer for the 1:100 dilution, followed by a 1:10 dilution series.
- Samples will be diluted further in the assay plate to yield 1,000 ng, 500 ng, 100 ng, and 10 ng (Figure 1).
- For best resolution, add 2 μl top Methylumbelliferone standard to 198 μl of stop buffer for the 1:100 dilution, followed by a 1:10 dilution series.
- Load the standard wells in triplicate to the flat bottom black 96-well plate as demonstrated in Figure 1.
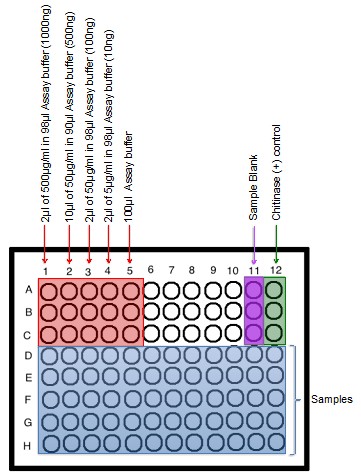
Figure 1. Plate set-up for chitinase assay. Red wells indicate 4-Methylumbelliferone standards, Purple indicates the sample blank (Working Solution), Green represents the positive control, and blue wells indicate sample wells.
- Dilute the chitinase positive control 1:200 in PBS to yield a final concentration of 1 μg/ml and load positive control wells with 10 μl chitinase and 90 μl working solution
- Use 100 μl working solution as the sample blank
- Add 90-99 μl working solution to each sample well in triplicate followed by 1-10 μl of each sample (samples are in lysis buffer)
- The amount of sample used for this assay will need to be optimized. Some samples may contain so much chitinase activity that the fluorescence will be saturated. This will vary considerably depending on the amount of cells plated and the chitinase activity of those cells.
- Wrap plate in foil and incubate at 37 °C for 30 min
- Incubation time may also need to be optimized. Cell lysates with high chitinase activity can be incubated for as little as 15 min. Alternatively, samples may be incubated for up to 1 h.
- Add 200 μl of stop buffer to each well to stop the reaction.
- Fluorescence can be measured on a plate reader at an excitation of 360 nm and emission of 450 nm within 30 min.
- Chitinase activity is calculated from the standard curve. Alternatively, chitinase activity may be calculated using the following equation:
Units/mL= (Fluoresencesample – Fluoresenceblank) x 1.9 x 0.3 x Dilution Factor
Fluoresence100 ng standard x reaction time x sample Volume
Recipes
- Assay buffer
Phosphate-Citrate Buffer pH=5.2 (26.7 ml of 0.2 M diabasic Sodium Phosphate, 23.3 ml of 0.1 M Citric Acid, top up to 100 ml DI water)
- Stop buffer
Glycine-NaOH buffer pH = 10.6 (combine 25 ml 0.2 M glycine stock solution with 22.75 ml 0.2 M NaOH, and dilute with DI water to make a 100 ml solution)
- Protein lysis buffer
50 mM Tris HCl (pH 7.5)
200 mM NaCl
10% Glycerol
0.5% TX-100
1 mM DTT (added to buffer fresh, just before adding to cultures)
- Substrate Stock
Prepare 20 mg/ml 4-Methylumbelliferyl substrate in DMSO.
- Positive control
Prepare 0.2 mg/ml chitinase from Trichoderma in PBS.
Acknowledgments
This protocol is adapted from Nance et al. (2012).
References
- Nance, J. P., Vannella, K. M., Worth, D., David, C., Carter, D., Noor, S., Hubeau, C., Fitz, L., Lane, T. E., Wynn, T. A. and Wilson, E. H. (2012). Chitinase dependent control of protozoan cyst burden in the brain. PLoS Pathog 8(11): e1002990.
Article Information
Copyright
© 2013 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Worth, D., Nance, J. P. and Wilson, E. H. (2013). Chitinase Assay from Cultured Bone Marrow Derived Macrophages. Bio-protocol 3(23): e983. DOI: 10.21769/BioProtoc.983.
- Nance, J. P., Vannella, K. M., Worth, D., David, C., Carter, D., Noor, S., Hubeau, C., Fitz, L., Lane, T. E., Wynn, T. A. and Wilson, E. H. (2012). Chitinase dependent control of protozoan cyst burden in the brain. PLoS Pathog 8(11): e1002990.
Category
Immunology > Immune cell function > Macrophage
Biochemistry > Protein > Activity
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










