- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Cell Fractionation of Pseudomonas aeruginosa
Published: Vol 3, Iss 19, Oct 5, 2013 DOI: 10.21769/BioProtoc.922 Views: 13755
Reviewed by: Fanglian He

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Prokaryotic Expression and Purification of the hSox2-HMG Domain
Lijie Yang [...] Jingjun Hong
Aug 20, 2025 2376 Views

Preparation and Negative Staining for Visualization of Cyanoglobule Lipid Droplets Using Transmission Electron Microscopy
Febri A. Susanto [...] Peter K. Lundquist
Dec 5, 2025 1311 Views

On-Column Dual-Gradient Refolding for Efficient Recovery of Insoluble Affinity-Tagged Recombinant Proteins
Anna Vlaskina [...] Maxim Patrushev
Feb 5, 2026 66 Views
Abstract
Pseudomonas aeruginosa is a Gram negative bacterium. Separating the cell envelope compartments enables proteins to be localized to confirm where in the cell they function. Cell fractionation can also provide a first step in a protein purification strategy (Williams et al., 1998). This protocol has been designed to obtain the different fractions of P. aeruginosa, namely the inner membrane, outer membrane, cytoplasmic and periplasmic compartments. Specific detection of the arginine specific autotransporter (AaaA) (Luckett et al., 2012) in the outer membrane of P. aeruginosa has been performed using this protocol.
Materials and Reagents
- P. aeruginosa strain (In this experiment, AaaA deficient mutant strains are used). The strain was bearing the pME6032 shuttle expression vector (Heeb et al., 2002). This vector is IPTG-inducible and has very good stability in Pseudomonas.
- Polyclonal antibody anti-AaaA (rabbit) (not commercial available) (Luckett et al., 2012)
- Anti-Rabbit IgG (whole molecule)–Peroxidase antibody produced in goat (Sigma-Aldrich, catalog number: A6154 )
- Isopropyl β-D-1-thiogalactopyranoside (IPTG) (Sigma-Aldrich, catalog number: I5502 )
- Ethylenediaminetetraacetic acid (EDTA) (Sigma-Aldrich, catalog number: EDS )
- Phenylmethanesulfonyl fluoride (PMSF) (Sigma-Aldrich, catalog number: P7626 )
- Luria Bertani (LB) (Difco, catalog number: 244620 )
- Autoclaved sterilized Phosphate buffered saline (PBS) (Sigma-Aldrich, catalog number: P4417 )
- Sucrose (Sigma-Aldrich, catalog number: S7903 )
- Sodium Lauryl Sarcoscine (SLS) (Sigma-Aldrich, catalog number: L5125 )
- Magnesium Sulphate (Sigma-Aldrich, catalog number: 63139 )
- MgCl2 (Sigma-Aldrich, catalog number: M2393 )
- DNAase I (Sigma-Aldrich, catalog number: D4527 )
- RNAase A (Sigma-Aldrich, catalog number: R5503 )
- 100% Trichloroacetic acid (TCA) (Sigma-Aldrich, catalog number: T6399 )
- Acetone (Sigma-Aldrich, catalog number: 179124 )
- NaOH (Sigma-Aldrich, catalog number: 221465 )
- Tris(hydroxymethyl) aminomethane (Sigma-Aldrich, catalog number: 252859 )
- Dithiothreitol (DTT) (Sigma-Aldrich, catalog number: D9779 )
- Sodium Dodecyl Sulphate (SDS) (Sigma-Aldrich, catalog number: L4509 )
- Bromophenol Blue (Sigma-Aldrich, catalog number: B0126 )
- Glycerol (Sigma-Aldrich, catalog number: G8773 )
- 4x Loading buffer (see Recipes)
Equipment
- French Pressure cell (Manufactured by Amicon and supplied by Thermo Fisher Scientific)
- 50 ml Falcon tube (BD Biosciences, Falcon®, catalog number: 352070 ) (Supplied by Scientific Laboratory Supplies)
- Eppendorf® Safe-Lock micro test tubes (Eppendorf, catalog number: 0030121023 ) (Supplied by Scientific Laboratory Supplies)
- Microfuge, larger volume centrifuge and high speed centrifuge (Beckman Coulter, model: Allegra X-22R Centrifuge )
- 1 ml pipettman
- W380 sonicator (Ultrasonics)
Procedure
- Set up overnight cultures for P. aeruginosa strains of interest from a single colony. Conditions: 10 ml LB broth in 50 ml Falcon tube at 37 °C shaking at 200 rpm.
- Next day, set up fresh cultures from the overnight cultures in new LB broth using a 1:100 ratio dilution, and grow at 37 °C shaking at 200 rpm until the exponential growth phase (0.4 - 0.5 OD600 nm).
- When the exponential phase is reached, add IPTG (1 mM final concentration) for 1 h to induce the production of the protein of interest if required.
- After the hour of induction, centrifuge the culture at 3,000 x g for 5 min at 4 °C. Discard the supernatant and gently resuspend the pellet in 10 ml of room temperature PBS using a 1 ml pipettman.
- Repeat step 4 twice so that the cells are washed three times in total.
- Resuspend the final pellet in 10 ml PBS solution.
- Check OD600 nm and dilute to an OD600 nm of 1.0 with PBS solution. This will be referred to as the ‘main solution’.
- Take 1 ml of the main solution to prepare the whole cell fraction. Centrifuge at 3,700 x g for 5 min in an Eppendorf tube and resuspend in 200 μl of 1x loading buffer, then sonicate for 10 seconds in W380 sonicator using the following settings: duty cycle, 40%; output control, 41/2 position and cycle time, continuous and boil at 100 °C for 10 minutes. The supernatant from the centrifugation can be kept on ice as a sample from the supernatant if secreted proteins are also to be analyzed.
- Take another 1 ml of the main solution to prepare the periplasmic and cytoplasmic fractions. Centrifuge at 3,700 x g for 2 min at room temperature in an Eppendorf tube.
- Wash with 300 μl of 25 mM Tris pH 7.4 three times by centrifuging at 3,700 x g for 5 min (room temperature) and gently resuspending the pellet with a pipettman.
- After the last centrifugation, the pellet needs to be resuspended in 50 μl of 25 mM Tris pH 7.4, 1 μl of 0.1 M EDTA and 50 μl of 40% w/w sucrose in 25 mM Tris pH 7.4.
- Mix the sample gently at room temperature for 10 min.
- Centrifuge at 3,700 x g for 5 min (room temperature), discard supernatant and resuspend pellet in 100 μl of ice cold 0.5 mM Magnesium Sulphate.
- Incubate on ice for 10 min, gently inverting occasionally to mix.
- Centrifuge for 5 min at 10,300 x g in microfuge (4 °C).
- The supernatant of the sample has the periplasmic fraction, and should be stored on ice.
- The pellet needs to be resuspended in 600 μl of 10 mM Tris pH 7.4 containing 20 μg/ml PMSF.
- The sample from step 17 should be frozen and thawed three times on dry ice.
- Add 19.9 μl MgCl2 (1 M) and 1.2 μl DNase I (1 mg/ml) to the sample from step 18. Mix by inversion.
- Incubate at 37 °C for 15 min.
- Centrifuge for 15 min at 10,300 x g, and the supernatant contains the cytoplasmic fraction, which should be stored on ice.
- The rest of main solution (approximately 5 ml) is used to get the membrane fractions.
- Centrifuge at 3,000 x g for 10 min at 4 °C, discard supernatant and resuspend pellet in 3 ml 20 mM Tris pH 7.4 containing 0.1 mg/ml DNase and 0.1 mg/ml RNase.
- Pass through the French Press three times at 16,000 lb/in, on ice.
- Centrifuge 3,000 x g for 20 min at 4 °C to remove unlysed cells.
- The resultant supernatant is transferred to a fresh tube, and centrifuged at 30,000 x g for 40 minutes at 4 °C.
- Discard the supernatant and resuspend the pellet in 200 μl 20 mM Tris pH 7.4 containing 0.7% (w/v) SLS.
- Incubate at 4 °C for 25 minutes and centrifuge at 30,000 x g for 40 minutes at 4 °C.
- The supernatant contains the inner membrane fraction, and should be stored in a fresh tube on ice.
- The pellet needs to be resuspended in 200 μl 20 mM Tris pH 7.4 and contains the outer membrane.
Note: The samples (periplasmic, inner membrane, cytoplasmic outer membrane fractions and culture supernatant) need to be subjected to TCA precipitation (Cooksley et al., 2003) to concentrate the proteins and remove residual detergents and salts. To TCA precipitate the proteins add TCA (from a 100% stock solution kept at 4 °C in the dark) to a final concentration of 10% v/v. Incubate the samples on ice for 30 minutes and centrifuge for 15 minutes at top speed in a microfuge at room temperature or 4 °C. Discard the supernatant carefully without disturbing the small and fragile pellet. Add 500 μl of ice cold acetone to the pellet, and centrifuge for 5 minutes in the microfuge. The supernatant is discarded again and the pellet air dried for 15 min. Finally, the pellet is resuspended in 20 μl of 50 mM NaOH plus 180 μl loading buffer 1x, sonicated for 10 sec, and boiled for 10 min. - The proteins can then be visualized by SDS-PAGE or Immunoblotting (Cooksley et al., 2003) as seen in Figure 1. It is good practice to have available control antibodies against proteins known to be localized to each of the cell fractions analyzed to verify that there is no cross contamination.
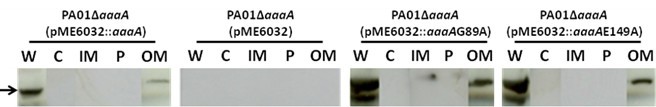
Figure 1. AaaA is found in the outer membrane fraction of P. aeruginosa. Cell fractionation was performed as described, followed by immunoblotting with specific anti-AaaA sera as described previously (Luckett et al., 2012). Whole cell (W), cytoplasmic (C), Inner Membrane (IM), Perplasmic (P) and outer membrane (OM) fractions are shown, and the expected position of AaaA is indicated by the arrow. The P. aeruginosa strains analyzed were the AaaA deficient mutant containing the plasmid pME6032 [PA01 delta aaaA (pME6032)], or a derivative of pME6032 with an insert encoding wild type AaaA [PA01 delta aaaA (pME6032::aaaA)] or a site directed mutant of AaaA replacing alanine with the residue at either position G89 [PA01 delta aaaA (pME6032:: aaaA G89A)] or E149 [PA01 delta aaaA (pME6032::aaaA E149A)]. To verify that the fractionation has occurred correctly, it is important to strip the blots and reprobe with antibodies specific for proteins that are only found in the fractions of interest as was shown in Luckett et al. (2012).
Recipes
- 4x Loading buffer
200 mM Tris(hydroxymethyl) aminomethane (pH 6.8)
800 mM Dithiothreitol (DTT)
8% Sodium Dodecyl Sulphate (SDS)
0.4% w/v Bromophenol Blue
40% v/v Glycerol
Acknowledgments
We acknowledge the use of this protocol in Luckett et al. (2012). We would like to thank the Chilean Government for financially supporting Esteban Paredes. We also thank Prof Miguel Camara for critical analysis of our work and everyone else in the Bacteriology Laboratories of the Centre of Biomolecular Sciences, University of Nottingham for helpful discussion about our work.
References
- Cooksley, C., Jenks, P. J., Green, A., Cockayne, A., Logan, R. P. and Hardie, K. R. (2003). NapA protects Helicobacter pylori from oxidative stress damage, and its production is influenced by the ferric uptake regulator. J Med Microbiol 52(Pt 6): 461-469.
- Heeb, S., Blumer, C. and Hass, D. (2002). Regulatory RNA as mediator in GacA/RsmA-dependent global control of exoproduct formation in Pseudomonas fluorescent CHA0. J Bacteriol 184(4):1046-56.
- Methods in Microbiology: Bacterial Pathogenesis, Academic Press. (1998). Eds: Paul Williams, Peter Williams, George Salmond, Julian Ketley Chapter 6.1: p185-191.
- Luckett, J. C., Darch, O., Watters, C., Abuoun, M., Wright, V., Paredes-Osses, E., Ward, J., Goto, H., Heeb, S., Pommier, S., Rumbaugh, K. P., Camara, M. and Hardie, K. R. (2012). A novel virulence strategy for Pseudomonas aeruginosa mediated by an autotransporter with arginine-specific aminopeptidase activity. PLoS Pathog 8(8): e1002854.
Article Information
Copyright
© 2013 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Paredes-Osses, E. and Hardie, K. R. (2013). Cell Fractionation of Pseudomonas aeruginosa. Bio-protocol 3(19): e922. DOI: 10.21769/BioProtoc.922.
- Luckett, J. C., Darch, O., Watters, C., Abuoun, M., Wright, V., Paredes-Osses, E., Ward, J., Goto, H., Heeb, S., Pommier, S., Rumbaugh, K. P., Camara, M. and Hardie, K. R. (2012). A novel virulence strategy for Pseudomonas aeruginosa mediated by an autotransporter with arginine-specific aminopeptidase activity. PLoS Pathog 8(8): e1002854.
Category
Microbiology > Microbial cell biology > Organelle isolation
Cell Biology > Organelle isolation > Fractionation
Biochemistry > Protein > Isolation and purification
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link