- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
RNA Isolation From Meloidogyne Spp. Galls
Published: Vol 3, Iss 17, Sep 5, 2013 DOI: 10.21769/BioProtoc.879 Views: 10048
Reviewed by: Tie Liu

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Simple Method for Efficient RNA Extraction From Arabidopsis Embryos
Fernanda Marchetti [...] Eduardo Zabaleta
Feb 20, 2025 1926 Views

An Optimized RNA Extraction Method From Micro-quantities of Guinea Pig Cartilage and Synovium for Osteoarthritis Research
Nidhi Bhardwaj [...] Jyotdeep Kaur
Jun 20, 2025 1618 Views

A Comparative Protocol for Preserving Deep-Water Marine Invertebrate Tissues: DNA/RNA Shield vs. Liquid Nitrogen for Dual Extraction of High-Quality Nucleic Acids
Ana S. Gomes [...] Olivier Laroche
Nov 20, 2025 1335 Views
Abstract
We describe an efficient method to obtain a sufficient quantity of RNA from nematode-induced galls with a high quality and integrity, proved to be appropriate for transcriptomic analysis, i.e. real time PCR, microarray hybridization or second generation sequencing. This protocol is efficient for small quantities of galls (organs with high protein and sugar contents). The protocol allows obtaining an RNA yield of 5-15 μg total RNA from 250-300 hand dissected galls at 3 days post infection (dpi) (Figure 1). It was proved particularly for Arabidopsis and tomato.
Keywords: Galls 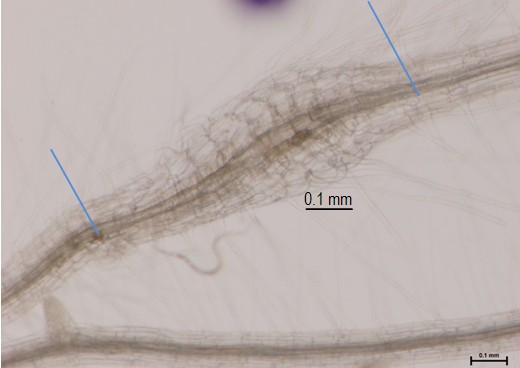
Figure 1. 3 dpi Arabidopsis gall from M. javanica. Blue lines indicate the collected material, galls with a small portion of roots (that allows easy handling), frozen and processed for RNA extraction.
Materials and Reagents
- Arabidopsis infected by M. javanica (3 dpi or 7 dpi)
- TRI Reagent (Molecular Research Centre, MRC, catalog number: TR-118 )
- Chloroform (Scharlau, catalog number: CL02031000 )
- Isopropanol (Merck KGaA, catalog number: 0 9634 )
- Sodium citrate (Sigma-Aldrich, catalog number: s4641 )
- Sodium chloride (Duchefa, catalog number: S0520 )
- NaOH (Duchefa, catalog number: s0523 )
- Ethanol (Merck KGaA, catalog number: 100986 )
- Diethylpyrocarbonate (DEPC) (Sigma-Aldrich, catalog number: D5758 )
- RNase-free water
- Acetone (Thermo Fisher Scientific, catalog number: A/0600/17 )
- Liquid nitrogen
- RNeasy Mini Kit (QIAGEN, catalog number: 74104 ))
- RNase-Free DNase Set (QIAGEN, catalog number: 79254 )
- High salt precipitation solution (see Recipes)
Equipment
- Centrifuges
- Microfuge
- 1.5 ml Eppendorf® tubes
- Porcelain mortar and pestles, textured surface on bowl interior, 60 mm diameter
- Fume hood
Procedure
Note: Use gloves and goggles for all procedure.
- Gall collection: To perform the RNA isolation from galls with a good efficiency, 250-300 hand dissected galls at 3 days post infection (3 dpi) or 100-150 galls at 7 dpi, should be collected. Collect galls and rapidly freeze them to a 1.5 ml Eppendorf® tube in liquid nitrogen.
Note: If scaled down, the RNA yield is not proportional to the amount of tissue. You can accumulate galls from independent collection events in the same Eppendorf®.
- Store at -80 °C until further processing (step C).
- Homogenization:
- Clean mortar and pestles with acetone, let them dry on the bench and autoclaved to avoid RNase activity. CAUTION: Check that mortar, pestle and pipettes used during the extraction have not been in contact with RNases. Sometimes it is preferable to keep material exclusively for RNA extraction.
- Add liquid nitrogen into the mortar with the pestle to cold them down.
- Add the galls into the mortar and proceed into a fume hood.
- When liquid nitrogen is nearly evaporated, add TRI Reagent® (500-750 μl/200-300 galls at 3 dpi); the total volume should not exceed 10% of the volume of TRI Reagent used for homogenization, as suggested by the TRI Reagent® protocol.
- Grind galls with a mortar and pestle. TRI Reagent solution initially becomes frozen, but it will melt gradually as a result of the homogenization. Proceed until a liquid appearance is observed and no visible tissue pieces are identified within the Reagent solution.
Note: Do not leave traces of galls without homogenizing, since this step can be limiting for good efficiency.
- Transfer the gall homogenate with a pipette to a 1.5 ml Eppendorf® tube and place it on ice while you homogenize the rest of the samples.
- Clean mortar and pestles with acetone, let them dry on the bench and autoclaved to avoid RNase activity. CAUTION: Check that mortar, pestle and pipettes used during the extraction have not been in contact with RNases. Sometimes it is preferable to keep material exclusively for RNA extraction.
- Phase separation
- Centrifuge at 12,000 x g for 10 min at 4 °C and transfer the supernatant containing the RNA to a new 1.5 ml Eppendorf® tube.
Note: The resulting pellet contains membranes, polysaccharides and high molecular weight DNA while the supernatant contains RNA. All insoluble material should be removed from the homogenate.
- Add 100 μl of chloroform per 500 μl of TRl Reagent® in a fume hood. Cap sample tubes securely.
- Shake tubes vigorously by hand for 15 s and incubate them at room temperature (RT) for 5 to 10 min.
Note: Do not use vortex.
- Centrifuge the samples at 12,000 x g for 15 min at 4 °C.
- Transfer the upper aqueous phase to a new tube.
- Centrifuge at 12,000 x g for 10 min at 4 °C and transfer the supernatant containing the RNA to a new 1.5 ml Eppendorf® tube.
- RNA precipitation
- Add 125 μl of isopropanol and 125 μl of high salt buffer per 500 μl of TRl Reagent®. Mix the solution and store it for 5-10 min at RT.
Note: The high salt precipitation solution removes contaminating compounds as proteoglycan and polysaccharide from the isolated RNA.
- Centrifuge at 12,000 x g for 8 min at 4 °C.
- Discard the supernatant.
- Add 125 μl of isopropanol and 125 μl of high salt buffer per 500 μl of TRl Reagent®. Mix the solution and store it for 5-10 min at RT.
- RNA Wash
- Wash the RNA pellet with 1 ml of 75% ethanol and mix by inversion several times.
- Centrifuge at 7,500 x g for 5 min at 4 °C and discard the supernatant.
- Repeat steps E-1 and 2.
- Spin 10 sec in a microfuge and discard carefully the remaining liquid.
- Air dry the pellet for 3-5 min. Do not let the RNA over-dry, as this will make it difficult to dissolve. The white RNA pellet will turn clear when it dries out.
- Add 100 μl DEPC-water or RNase-free water immediately after the pellet becomes clear.
- Dissolve RNA by incubating for 10 min at 37 °C followed by 5 min at 60 °C.
- Store RNA solution at -80 °C or continue with further RNA Cleanup by DNase digestion.
- Wash the RNA pellet with 1 ml of 75% ethanol and mix by inversion several times.
- Combined in column DNase I Digestion and RNA Cleanup. This procedure is based on the combined use of two different commercial kits (RNase-Free DNase Set and RNeasy Mini Kit). Please, use handling instructions of all buffers and reagents following their recommendations.
Note: Buffers as RPE, RTL, RW1, RDD are supplied by the companies within the kits. Before starting RNA cleanup, make sure the DNase I and Buffer RPE are ready to use.- Preparation of DNase I stock solution: Inject 550 μl RNase-free water into the lyophilized DNase I vial using an RNase-free needle and syringe. Mix gently by inverting the vial and divide it in 10 μl single-use aliquots. Store at -20 °C for up 9 months.
- Preparation of Buffer RPE: Add 4 volumes of 96-100% ethanol (44 ml) as indicated in the bottle.
- Adjust the RNA sample volume to 100 μl with RNase-free water.
- Add 350 μl of Buffer RLT and mix well by pipetting.
- Add 250 μl EtOH, mix well by pipetting.
- Transfer the sample to an RNeasy Mini spin column placed in a 2 ml collection tube. Close the lid gently and centrifuge at 8,000 x g for 15 sec. Discard the flow-through.
- Add 350 μl buffer RW1 to the RNeasy spin column. Close the lid gently and centrifuge at 8,000 x g for 15 sec. Discard the flow-through.
- Add 70 μl buffer RDD (supplied with the RNase-Free DNase Set) to 10 μl DNase I stock solution. Mix gently by inverting the tube.
Note: Do not vortex.
- Add 80 μl of the DNase I incubation Mix to the RNeasy spin column, and place on the bench at RT for 15 min.
Note: Add the Mix directly to the center of the RNeasy spin column membrane.
- Add 350 μl buffer RW1 to the RNeasy spin column. Close the lid gently and centrifuge at 8,000 x g for 15 sec. Discard the flow-through.
- Add 500 μl buffer RPE to the RNeasy spin column. Close the lid gently and centrifuge at 8,000 x g for 15 sec. Discard the flow-through.
- Add 500 μl buffer RPE to the RNeasy spin column. Close the lid gently and centrifuge at 8,000 x g for 2 min.
Note: The long centrifugation dries the spin column membrane, eliminating ethanol contamination.
- Quickly remove the RNeasy spin column from the collection tube so that the column does not contact the flow through.
- Place the RNeasy spin column in a new 2 ml collection tube. Close the lid gently and centrifuge at full speed for 1 min.
Note: It is important to eliminate any possible buffer RPE carryover.
- Place the RNeasy spin column in a new 1.5 ml collection tube. Add 30 μl RNase-free water or DEPC-water to the center of the spin column membrane. Incubate 1 min at RT. Close the lid gently and centrifuge at 8,000 x g for 1 min to elute the RNA.
- Add 20 μl RNase free water or DEPC-water to the center of spin column membrane to elute the maximum RNA. Incubate 1 min at RT. Close the lid gently and centrifuge at 8,000 x g for 1 min.
- Use Nanodrop and/or Bioanalyzer to test the RNA quantity and quality. You should get 5-10 μg total RNA from 250-300 collected galls at 3 dpi of a high quality.
- Store the RNA sample in -80 °C for future use.
- Preparation of DNase I stock solution: Inject 550 μl RNase-free water into the lyophilized DNase I vial using an RNase-free needle and syringe. Mix gently by inverting the vial and divide it in 10 μl single-use aliquots. Store at -20 °C for up 9 months.
Recipes
- High salt precipitation solution
0.8 M sodium citrate
1.2 M NaCl
Acknowledgments
This protocol is adapted from Barcala et al. (2010) and Portillo et al. (2013).
References
- Barcala, M., Garcia, A., Cabrera, J., Casson, S., Lindsey, K., Favery, B., Garcia-Casado, G., Solano, R., Fenoll, C. and Escobar, C. (2010). Early transcriptomic events in microdissected Arabidopsis nematode-induced giant cells. Plant J 61(4): 698-712.
- Portillo, M., Cabrera, J., Lindsey, K., Topping, J., Andres, M. F., Emiliozzi, M., Oliveros, J. C., Garcia-Casado, G., Solano, R., Koltai, H., Resnick, N., Fenoll, C. and Escobar, C. (2013). Distinct and conserved transcriptomic changes during nematode-induced giant cell development in tomato compared with Arabidopsis: a functional role for gene repression. New Phytol 197(4): 1276-1290.
- Portillo, M., Fenoll. C. and Escobar C. (2006). Evaluation of different RNA extraction methods for small quantities of plant tissue: Combined effects of reagent type and homogenisation procedure on RNA quality-integrity and yield. Physiol Plantarum 128 (1): 1-7.
Article Information
Copyright
© 2013 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
García, A., Barcala, M., Cabrera, J., Fenoll, C. and Escobar, C. (2013). RNA Isolation From Meloidogyne Spp. Galls. Bio-protocol 3(17): e879. DOI: 10.21769/BioProtoc.879.
Category
Plant Science > Plant physiology > Endosymbiosis
Molecular Biology > RNA > RNA extraction
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











