- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Sodium Current Measurements in HEK293 Cells
Published: Vol 3, Iss 16, Aug 20, 2013 DOI: 10.21769/BioProtoc.858 Views: 14603
Reviewed by: Anonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

A Protocol for Measurement of Intracellular pH
Iman Saramipoor Behbahan [...] Siavash K. Kurdistani
Jan 20, 2014 23483 Views

Analyzing the Quenchable Iron Pool in Murine Macrophages by Flow Cytometry
Michael Riedelberger and Karl Kuchler
Mar 20, 2020 6242 Views
Abstract
This protocol is used to measure sodium currents from heterologously transfected cells lines, such as HEK293 cells. Standard whole cell patch clamp technique is used to assess ion channel function. This protocol has been used to study cardiac type Nav1.5 sodium channel, and in particular to compare wild-type and mutant channels related to Brugada Syndrome (BrS). Mutations related to BrS provoke a loss of function of the cardiac sodium channel. This is evidenced by a reduction of sodium current density and/or alterations of the activation or inactivation kinetic parameters, such as a slow recovery from inactivation. These effects could explain the alteration of the cardiac action potential that leads to the characteristic ST elevation observed in the electrocardiogram of the Brugada Syndrome patients. This protocol, with some variations, is suitable for studying other cardiac arrhythmias related to alterations in the cardiac type sodium channel function as well as for studying other voltage-dependent sodium channels.
Materials and Reagents
- Human embryonic kidney HEK293 cells (Health Protection Agency Culture Collections, catalog number: 96121229 )
- Vector/s: mammalian expression vectors harboring the cDNA of interest (i.e. pcDNA3 + SCN5A)
- Dulbecco’s Modified Eagle’s Medium (DMEM) (Sigma-Aldrich, catalog number: D6546 )
- Fetal Bovine Serum (Sigma-Aldrich, catalog number: F4135 )
- Penicillin-streptavidin (Life Technologies, Gibco®, catalog number: 15140-122 )
- GlutaMAXTM (Life Technologies, Gibco®, catalog number: 35050-038 )
- 0.05% Trypsin-EDTA (Life Technologies, Gibco®, catalog number: 25300-062 )
- Dulbecco’s Phosphate Buffered Saline (Sigma-Aldrich, catalog number: D8662 )
- GeneCellinTM Transfection Reagent (BioCellChallenge, catalog number: GC-1000 )
- Opti-MEM® Reduced Serum Media + GlutaMAXTM (Life Technologies, Gibco®, catalog number: 31985-062 )
- Sticky wax in bars (Cera de Reus, catalog number: 25005001 )
- Sodium chloride (NaCl) (Sigma-Aldrich)
- Potassium chloride (KCl) (Sigma-Aldrich)
- Cesium chloride (CsCl) (Sigma-Aldrich)
- Calcium chloride (CaCl2) (Sigma-Aldrich)
- Magnesium chloride (MgCl2) (Sigma-Aldrich)
- N-2-hydroxyethylpiperazine-N’-2-ethanesulfonic acid (HEPES) (Sigma-Aldrich)
- Ethylene glycol-bis (2-amino-ethylether)-N,N,N’,N’-tetra-acetic acid (EGTA) (Sigma-Aldrich)
- Adenosine 5′-triphosphate magnesium salt (ATP-Mg2+) (Sigma-Aldrich)
- Sodium hydroxide (NaOH) (Sigma-Aldrich)
- Cesium hydroxide (CsOH) (Sigma-Aldrich)
- Glucose (Sigma-Aldrich)
- Bath solution (see Recipes)
- Pipette solution (see Recipes)
Equipment
- Inverted microscope Nikon Eclipse Ti fitted for epifluorescence (Nikon Instruments Inc.)
- Glass capillary PC-10 Puller (Narishige International USA Inc.)
- Micromanipulator MP-285 (Sutter Instrument Co.)
- Vapor pressure Osmometer VAPOR 5520 (Wescor Inc.)
- Axopatch 200B Capacitor Feedback Patch Clamp Amplifier (Molecular Devices)
- Axon Digidata 1440A Data Acquisition System (Molecular Devices)
- CO2 incubator
- Nunclon® cell culture dishes 35 x 10 mm (Sigma-Aldrich, catalog number: D7804 )
- Glass capillaries (Brand GmbH + CO KG, catalog number: 7493-21 )
Software
- pCLAMP 10.2 Electrophysiology Data Acquisition and Analysis Software (Molecular Devices)
- OriginPro 8 software (OriginLab Corporation)
Procedure
- Cell culture and transfection
- The day before transfection, plate HEK293 cells in 35 mm dishes at a density that will result in 70% confluence 24 h later (this is 2.1 x 105 cells but it may vary depending on how fast your cell line grows), and maintain in a CO2 incubator at 37 °C in DMEM supplemented with 10% Fetal Bovine Serum, 1% Penicillin-streptomycin and 1% GlutaMAXTM.
- Perform the transfection for each 35 mm dish of HEK293 cells with a mix of 200 μl Opti-MEM® Medium, 3 μg (in total) of the vectors of interest and 4 μl of GeneCellinTM Transfection Reagent. The inclusion of a vector harboring the cDNA encoding a fluorescent protein will allow the identification of transfected cells.
Note: Other transfection reagents and methods have been successfully used for transfecting sodium channels. - Twenty-four hours after transfection, wash the each dish with 1.5 ml DPBS, then add 0.2 ml 0.05% Trypsin-EDTA and incubate at 37 °C for 2 min, cells should look separated from each other, add 1.5 ml of DMEM to the dish, disperse with a fine tip pipette and plate 200-300 μl in a new dish with 1.5 ml of media.
Note: You should get individual cells for recording.
- The day before transfection, plate HEK293 cells in 35 mm dishes at a density that will result in 70% confluence 24 h later (this is 2.1 x 105 cells but it may vary depending on how fast your cell line grows), and maintain in a CO2 incubator at 37 °C in DMEM supplemented with 10% Fetal Bovine Serum, 1% Penicillin-streptomycin and 1% GlutaMAXTM.
- Patch-clamp procedure (48 h after transfection)
- Pull pipettes from glass capillaries and finely coat the tip with melted dental wax (be careful not to occlude the tip hole). Their resistance should range from 2 to 4 MΩ when filled with the Pipette solution.
- Remove DMEM from the transfected cells dish that you will use and, after rinsing the cells twice with the Bath solution (approximately 3 ml in total), fill it to about one third of the height with the same solution.
- Choose the cells that you will record from individual fluorescent cells.
- Fill the pipette with the pipette solution and place it in the electrode holder. Lower the pipette to place it in the Bath solution. After compensating offsets, approach the pipette to the chosen cell with the help of the remote micromanipulator to form a high resistance cell-attached seal.
- Once the seal is formed and the whole cell configuration is established, compensate series resistance at 80-90%.
- Wait five minutes before starting to record. This allows the cell content to equilibrate with the pipette solution.
- For acquisition, set your filter at 5 kHz and your sampling rate at 20-25 kHz.
- Pull pipettes from glass capillaries and finely coat the tip with melted dental wax (be careful not to occlude the tip hole). Their resistance should range from 2 to 4 MΩ when filled with the Pipette solution.
- Sodium measurements protocols
- Macroscopic sodium current: Currents are elicited by 50 ms depolarizing steps (from -80 to +80 mV in 5 mV increments) from a holding potential of -120 mV.
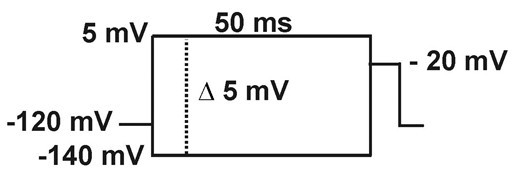
- Steady-state inactivation (h∞): Current is measured with -20 mV pulses (20 ms), following 50 ms pre-pulses to different potentials (-140 to +5 mV in 5 mV increments).
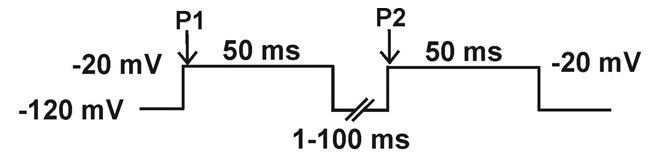
- Recovery from inactivation: current is elicited by a -20 mV, 20 ms, pulse (P2), preceded by a 50 ms depolarizing pre-pulse to -20 mV (P1) from a holding potential of -120 mV, followed by a hyperpolarizing pulse to -120 mV of increasing duration (1-100 ms).
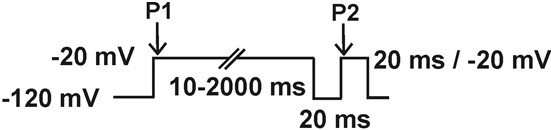
- Slow inactivation: Double pulse protocol 1: Current is measured with a test pulse to -20 mV during 20 ms (P2), preceded by a depolarizing pulse from -120 mV to -20 mV (P1) of increasing duration (10-100 ms in 10 ms increments), followed by a hyperpolarizing pulse to -120 mV during 20 ms, to remove fast inactivation. Double pulse protocol 2: This protocol is the same as the previous one except for that the duration of the first depolarizing pulse to -20 (P1) runs from 100 to 2,000 ms in 200 ms increments.
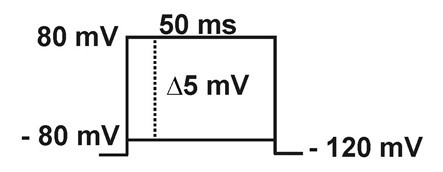
- Macroscopic sodium current: Currents are elicited by 50 ms depolarizing steps (from -80 to +80 mV in 5 mV increments) from a holding potential of -120 mV.
- Sodium measurements analysis (pClamp 10.2 and OriginPro 8 software)
- Current density-voltage (pA/pF): The measured peak current at the different voltages applied is normalized by the cell capacitance.
- Activation curve: Whole cell conductance (G) is obtained from the current- density relationship (I-V) by dividing the peak current obtained at each potential by the driving force (V-Eion) at each potential. These values are normalized to the maximum conductance (Gmax) and plotted against each voltage. Data is fitted to a Boltzmann equation of the form G=Gmax/ (1 + exp[(V1/2-V)/k]) where V is the applied potential, V1/2 is the voltage at which half of the channels are activated, and k is the slope factor.
- Inactivation time constants: time constants (t), tslow and tfast, are obtained from fitting the currents elicited with the macroscopic sodium current protocol to a second order exponential function. At some voltages, when the current is either too fast or too small, it is not possible to fit a second order exponential. In this case a first order exponential is used. The region analyzed to obtain the time constants is comprised from the peak of the current to a point where the current has reached a plateau near zero. tslow and tfast are plotted against voltage.
- Steady-state inactivation curve: Peak current amplitude (I) is normalized to the maximum peak current amplitude (Imax). The I/Imax values from the test pulse are plotted against the voltage during the pre-pulse. Experimental data is fitted to a Boltzmann equation of the form I = Imax/ (1 + exp[(V-V1/2)/k]), where V is the applied voltage, V1/2 is the voltage at which half of the channels are inactivated and k is the Boltzmman constant.
- Recovery from inactivation: Recovery current values are obtained by dividing the peak current from P2 by the peak current at P1. P2/P1 ratios are plotted against the recovery interval times. The recovery from inactivation curve is fitted to a mono-exponential function to obtain the time constants (t).
- Slow inactivation: Peak current is measured at P2 and P1. The P2/P1 ratio is plotted against the depolarizing pulse interval times. The slow inactivation curve is fitted to a mono-exponential function to obtain the slow inactivation constant (t).
- Current density-voltage (pA/pF): The measured peak current at the different voltages applied is normalized by the cell capacitance.
Recipes
- Bath solution
140 mM NaCl
3 mM KCl
10 mM HEPES
1.8 mM CaCl2
1.2 mM MgCl2
The pH is adjusted to 7.4 with NaOH
Osmolality is adjusted by the addition of glucose to approximately 325 mOsm. - Pipette solution
130 mM CsCl
1 mM EGTA
10 mM HEPES
10 mM NaCl
2 mM ATP-Mg2+
The pH is adjusted to 7.2 with CsOH
Osmolality is adjusted by the addition of glucose to approximately 308 mOsm.
Note: The difference in osmolality between Pipette and Bath solutions should be near 5%.
Acknowledgments
This protocol is adapted from Riuro et al. (2013) and Tarradas et al. (2013).
References
- Riuró, H., Beltran‐Alvarez, P., Tarradas, A., Selga, E., Campuzano, O., Vergés, M., Pagans, S.,Iglesias, A., Brugada, J. and Brugada, P. (2013). A Missense Mutation in the Sodium Channel β2 Subunit Reveals SCN2B as a New Candidate Gene for Brugada Syndrome. Hum Mutat 34(7):961-966.
- Tarradas, A., Selga, E., Beltran-Alvarez, P., Perez-Serra, A., Riuro, H., Pico, F., Iglesias, A., Campuzano, O., Castro-Urda, V., Fernandez-Lozano, I., Perez, G. J., Scornik, F. S. and Brugada, R. (2013). A novel missense mutation, I890T, in the pore region of cardiac sodium channel causes Brugada syndrome. PLoS One 8(1): e53220.
Additional suggested bibliography
- Sakmann, Bert and Neher, Erwin. (1995). Single-Channel Recording. Springer-Verlag, 2nd Edition. New York.
- Molleman, Areles (reprinted 2008). Patch Clamping. An Introductory guide to patch clamp electrophysiology. John Wiley and Sons Ltd. England: West Sussex.
Article Information
Copyright
© 2013 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Riuró, H., Tarradas, A., Selga, E., Brugada, R., Scornik, F. and Pérez, G. (2013). Sodium Current Measurements in HEK293 Cells. Bio-protocol 3(16): e858. DOI: 10.21769/BioProtoc.858.
Category
Cell Biology > Cell-based analysis > Ion analysis > Sodium
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









