- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Cell Surface Protein Biotinylation and Analysis
Published: Vol 3, Iss 16, Aug 20, 2013 DOI: 10.21769/BioProtoc.857 Views: 33715
Reviewed by: Anonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Protocol for the Preparation of a Recombinant Treacle Fragment for Liquid–Liquid Phase Separation (LLPS) Assays
Nadezhda V. Petrova [...] Artem K. Velichko
Sep 20, 2025 1823 Views

Whole-Mount Visualization of Primary Cilia in the Developing Mouse Brain
Oscar Torres Gutierrez [...] Xuecai Ge
Dec 20, 2025 1012 Views

Optimized Secretome Sample Preparation From High Volume Cell Culture Media for LC–MS/MS Proteomic Analysis
Basil Baby Mattamana [...] Peter Allen Faull
Dec 20, 2025 1261 Views
Abstract
A great way to specifically isolate and quantify proteins in the cell surface membrane is to take advantage of the biotinylation technique. It consists of labeling cell surface proteins with a biotin reagent before lysing the cells, and isolating these tagged proteins by NeutrAvidin pull-down. Then, the samples are subjected to SDS-PAGE separation, transferred to PVDF membranes and probed with specific antibodies. Quantification of cell surface expression is accomplished by densitometric measurement of the bands corresponding to the protein of interest and subsequent normalization by a membrane protein (as control).
Materials and Reagents
- Human embryonic kidney HEK293 cells (Health Protection Agency Culture Collections, catalog number: 96121229 )
- Biotin reagent: EZ-link Sulfo-NHS-LC-LC-biotin (Thermo-Pierce, catalog number: 21338 )
- Electrophoresis grade Glycine (Sigma-Aldrich, catalog number: G8898 )
- Immobilized/NeutrAvidin Ultralink Resin (Thermo-Pierce, catalog number: 53150 )
- PierceTM BCA Protein Assay Kit (Thermo-Pierce, catalog number: 23227 )
- Polyvinylidene difluoride (PVDF) membrane (GE Healthcare Life Sciences, catalog number: RPN303F )
- Nonfat dry milk (AppliChem GmbH, catalog number: A0830 )
- Chemiluminescence substrate: Super signal west femto maximum sensitivity substrate (Thermo Fisher Scientific, catalog number: 34096 )
- X-ray films: Amersham Hyperfilm ECL (GE Healthcare Life Sciences, catalog number: 28-9068-35 )
- Primary antibodies
- Secondary antibodies
- Protease inhibitors Cocktail (Roche Tablets, catalog number: 11 836 170 001 )
- DPBS with calcium and magnesium (DPBS+) (see Recipes)
- Lysis buffer LB1, LB2, LB3 (see Recipes)
- Phosphate-Buffered Saline (PBS) (see Recipes)
- Saline washing solution (SWS) (see Recipes)
- 3x Gel loading buffer (see Recipes)
- SDS-PAGE gel (see Recipes)
- 5x Running buffer (see Recipes)
- 10x Western blot transfer buffer (see Recipes)
Equipment
- 35 mm Dish (Thermo Fisher Scientific, catalog number: 153066 )
- 6-well cell culture plate (BD Biosciences, catalog number: 353046 )
- Cell scrapers (VWR International, catalog number: 734-2603 )
- 96-well Microtest Plate (Sarstedt, catalog number: 82.1581 )
- OrbitTM LS Low Speed Shaker (LABNET, catalog number: S-2030-LS )
- Rotating wheel (Noria R NR50, Ovan, catalog number: 10000-00062 )
- Microcentrifuge R5415 (Eppendorf, catalog number: 022621425 )
- Digital heat block (VWR International, catalog number: 460-3267 )
- Microplate Reader Benchmark plus (Bio-Rad Laboratories, catalog number: 170-6936 )
- Mini-PROTEAN Tetra Cell (Bio-Rad Laboratories, catalog number: 165-8000 )
- Mini Trans-Blot Module (Bio-Rad Laboratories, catalog number: 170-3935 )
- PowerPac HC Power Supply (Bio-Rad Laboratories, catalog number: 164-5052 )
Software
- ImageJ software (available at http://rsb.info.nih.gov/ij ) (National Institute of Health, NIH)
Procedure
- Biotinylation
Note: Perform all incubations on ice.- Remove growth media from the cells that you want to study, cultured in 35 mm plates.
Note: We used Human Embryonic Kidney HEK293 cells as experimental model. Cells were maintained in Dulbecco's Modified Eagle's Medium supplemented with 10% fetal bovine serum, 1% Penicilin-streptomycin and 1% Glumtamax at 37 °C and 5% CO2.
We plated approximately 2.1 x 105 HEK293 cells in 35 mm dishes, but this value may vary depending on how fast your cell line grows (this will result in 70% of confluence 24 h later, which is the day of the transfection). Forty-eight hours after transfection we performed the Cell Surface Protein Biotinylation protocol. - Wash cells twice with 1 ml of ice cold DPBS+.
- Incubate cells for 30 min on ice in the cold room with gentle rocking with 400 μl per plate of the biotin solution (2.5 mg/ml biotin reagent in DPBS+).
- Wash each plate for 5 min three times with 1 ml of cold 100 mM Glycine in DPBS+, on ice in the cold room with gentle rocking.
- Wash each plate for 5 min twice with 1 ml of cold 20 mM Glycine in DPBS+, as above.
- Lyse with 200 μl of lysis buffer LB3 into each plate and use cell scrapers to detach the cells. Collect the cell lysates in 1.5 ml tubes.
- For lysis, place the tubes in a rotating wheel at slow speed 1 h in the cold room.
- While the lysis is performed, prepare the Immobilized/ NeutrAvidin Ultralink beads: For each sample, take 40 μl of NeutrAvidin beads at 50% slurry and wash the beads twice with 0.5 ml DPBS+ and twice again with 0.5 ml LB2 (for washing, centrifuge 30 sec at 3,000 x g at 4 °C and remove the supernatant). After the last wash, resuspend the precipitated beads with 20 μl of LB3.
- Spin lysates at 16,000 x g (maximum speed) for 15 min at 4 °C.
- Transfer the supernatants (solubilized material) to 1.5 ml tubes. Keep 10-15% of each supernatant in another tube and store at -80 °C (these are the INPUT samples, and the rest of the supernatant will be referred to as PULL-DOWN samples).
- Incubate the pull-down samples with the 40 μl of NeutrAvidin beads prepared in step 8 overnight in the rotating wheel at slow speed in the cold room.
- Centrifuge the samples at 16,000 x g, 30 sec at 4 °C.
- Wash beads once with LB3, twice with LB2, twice with SWS and once with LB1. Each time, add 1 ml of the appropriate solution and centrifuge at 16,000 x g, 30 sec at 4 °C.
- Resuspend the precipitated proteins in 25 μl of 2x gel loading buffer.
- Heat the samples at 70 °C for 10 min in the heat block.
- Keep pull-down samples at -20 °C until SDS-PAGE and Western blot.
- Remove growth media from the cells that you want to study, cultured in 35 mm plates.
- Western blot
- Use part of the input samples to quantify the protein using the BCA Protein Assay Reagent alongside BSA standards, following the manufacturer's directions. From each input sample, transfer equal amounts of protein to a new tube and mix with 3x gel loading buffer. Heat the samples at 70 °C for 10 min.
Note: Other protein quantification methods can be used and are equally efficient. - Load in an SDS-PAGE gel the aliquots of the inputs prepared in step B-1 and the total volume of the pull-down samples (without the beads). Before loading, spin down the samples to pellet the beads.
- Run the gel at 40 V until samples have run through the stacking gel (30 min approximately). At that point, increase voltage to 100 V until the blue dye front has nearly run out of the gel.
- Prepare the sandwich: Soak the gel 15 min shaking in Western blot transfer buffer. Activate the Polyvinylidene difluoride (PVDF) membrane by soaking it for 10 sec with methanol, then twice with water and 10 min with Western blot Transfer Buffer. Also, wet two sheets of filter paper and two fiber pads with Western blot Transfer Buffer. Construct the sandwich with the gel on the cathode side and the membrane on the anode side, with a sheet of filter paper and a fiber pad at each side.
- Transfer the proteins to the PVDF membrane at 80 V for 2 h in the cold room.
- Remove the membrane from the sandwich and block it with 5% nonfat milk in PBS with 0.1% Tween-20 1 h at room temperature.
Note: All Western blot incubations were performed in a shaker at low speed. - Incubate the membrane with primary antibody overnight at 4 °C.
Note: We used antibodies against human Nav1.5 (protein of interest; rabbit polyclonal, at 1:1,000 in 5% non-fat milk in PBS with 0.1% Tween-20) and Na+/K+ ATPase (membrane protein used as control, mouse, at 1:5,000 in 5% non-fat milk in PBS with 0.1% Tween-20). - Wash three times with PBS with 0.1% Tween-20 to completely cover the membrane (approximately 20 ml), 10 min each, at room temperature.
- Incubate with the appropriate secondary antibody 1 h at room temperature.
Note: We used peroxidase conjugated- goat anti-rabbit or anti-mouse IgG antibodies for Nav1.5 and Na+/K+ ATPase, respectively (at 1:2,000 in 5% non-fat milk-PBS with 0.1% Tween-20). - Wash twice with PBS with 0.1% Tween-20 to completely cover the membrane (approximately 20 ml), 10 min each, at room temperature.
- For signal development, use the chemiluminescence substrate and follow the manufacturer's instructions. Remove excess reagent and cover the membrane in transparent plastic wrap.
- Detect the protein of interest by exposure to X-ray films (Figure 1).
Note: For normalization purposes, a membrane protein should also be assessed as control.
Figure 1. Western Blot detection of Nav1.5 and Na+/K+ ATPase proteins performed after cell surface biotinylation from WT and mutant cells
- Use part of the input samples to quantify the protein using the BCA Protein Assay Reagent alongside BSA standards, following the manufacturer's directions. From each input sample, transfer equal amounts of protein to a new tube and mix with 3x gel loading buffer. Heat the samples at 70 °C for 10 min.
- Quantification
- Scan the films in high resolution using the transparency mode.
- Use the ImageJ software for band quantification. Follow the user's guide instructions to determine intensity values for each band as the integrated density (sum of pixel values) within a fixed area.
Note: When the aim is to compare WT and mutant conditions within multiple replicates, proceed as follows:- Determine intensity values for the bands corresponding to WT and mutant conditions for the membrane protein used as control (ICWT and ICMUT, respectively) and WT and mutant condition for the protein of interest (IWT and IMut, respectively).
- Calculate the ratio between ICWT and ICMUT to obtain the normalization factor (Nf):ICWT/ICMUT = Nf
- Multiply IMut by the normalization factor calculated in the previous step to obtain the corrected mutant condition intensity value (IC Mut):IMut x Nf = IC Mut
- Divide IC Mut by IWT to obtain the expression of the mutant condition relative to IWT. IC Mut/IWT = mutant relative expression
- Calculate the average of the mutant relative expression values for all the replicates and perform statistical analysis to assess possible differences with respect to the WT (Figure 2).
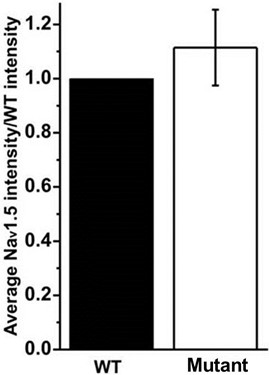
Figure 2. Bar graph shows the average of the intensity values obtained for each condition
- Determine intensity values for the bands corresponding to WT and mutant conditions for the membrane protein used as control (ICWT and ICMUT, respectively) and WT and mutant condition for the protein of interest (IWT and IMut, respectively).
- Scan the films in high resolution using the transparency mode.
Recipes
Note: All chemical reagents were obtained from Sigma-Aldrich unless stated otherwise.
- DPBS+ (For 2 L)
0.2 g of CaCl2
0.4 g KCl
0.4 g KH2PO4
0.2 g MgCl2.6H2O
16 g NaCl
4.32 g Na2HPO4.7H2O
To avoid forming an insoluble precipitate, add everything except CaCl2, and have it stirring in the cold room close to correct volume. While stirring and when cold, add CaCl2, and continue stirring for at least 10 min until the solution becomes clear. Add milli-Q water to the final volume of 2 L. - LB1
50 mM Tris/HCl (pH 7.4)
150 mM NaCl
1 mM EDTA - LB2
LB1 plus 1% (w/v) Triton X-100
To mix well, place the solution 1 h in cold room in the rotating wheel at slow speed. - LB3
For every 10 ml of solution LB2, add 1 tablet of protease inhibitors Cocktail right before use. - 1x PBS
8 g NaCl
0.2 g KCl
1.44 g Na2HPO4
0.24 g KH2PO4
Add mili-Q water for bring the final volume to 1 L. - SWS
0.1% Triton X-100 in PBS (pH 7.4)
350 mM NaCl
5 mM EDTA - 3x gel loading buffer
180 mM Tris/HCl (pH 6.8)
7.5% SDS
30% glycerol
0.051% Bromophenol blue
150 mM DTT - SDS-PAGE gel (for 2 gels 1.5 mm thick)
- Stacking (4%)
4.65 ml H2O
1.88 ml 0.5 M Tris (pH 7.4)
0.75 ml Acrylamide: Bisacrylamide 30% solution 37.5:1
75 μl 10% SDS
45 μl 10% Ammonium persulfate
15 μl N, N, N', N'-tetramethylethylenediamine (TEMED) - Resolving (4%)
9.35 ml H2O
3.75 ml 1.5 M Tris (pH 8.8)
1.46 ml Acrylamide: 30% Bisacrylamide solution 37.5:1
150 μl 10% SDS
69 μl Ammonium persulfate 10%
23.1 μl N, N, N', N'-tetramethylethylenediamine (TEMED)
- Stacking (4%)
- 5x Running buffer (For 1 L)
5 g SDS
144 g glycine
30 g Tris - 10x Western blot transfer buffer (For 1 L)
10 g SDS
24.24 g Tris
111.75 g glycine
Before use, make up 1x and add 20% Methanol.
Acknowledgments
The protocol was used in: Riuró et al. (2013); and Tarradas et al. (2013), but was adapted from a previously published paper: Cuartero et al. (2012). Funding sources at the time the protocol was developed included a grant from the Spanish Ministerio de Sanidad y Consumo to M. Verges (PI07/0895); Fellowship from the Príncipe Felipe Research Center (CIPF) to Y. Cuartero (PR 01/2007); and Ramón y Cajal contract from the Spanish Ministerio de Educación y Ciencia to M. Verges. Funding when it was implemented imcluded “La Caixa” Foundation to R. Brugada; Centro Nacional de Investigaciones Cardiovasculares (CNIC) Translational to R. Brugada (CNIC-03-2008); Ministerio de Sanidad y Consumo to R. Brugada (PI08/1800); Ministerio de Sanidad y Consumo fellowships or contracts (FI09/00336, CD09/00055, CD10/00275, CD11/00063 and PI2008/1800); Univ. of Girona fellowships to H. Riuró (BR2012/47); Ministerio de Sanidad y Consumo: Red Cooperativa de Insuficiencia Cardiaca (REDINSCOR) RD06/03/0018; and Sociedad Española de Cardiología (2011, Investigación Básica).
References
- Cuartero, Y., Mellado, M., Capell, A., Alvarez-Dolado, M. and Verges, M. (2012). Retromer regulates postendocytic sorting of beta-secretase in polarized Madin-Darby canine kidney cells. Traffic 13(10): 1393-1410.
- Riuró, H., Beltran‐Alvarez, P., Tarradas, A., Selga, E., Campuzano, O., Vergés, M., Pagans, S.,Iglesias, A., Brugada, J. and Brugada, P. (2013). A missense mutation in the sodium channel β2 subunit reveals SCN2B as a new candidate gene for brugada syndrome. Hum Mutat 34(7):961-966.
- Tarradas, A., Selga, E., Beltran-Alvarez, P., Perez-Serra, A., Riuro, H., Pico, F., Iglesias, A.,Campuzano, O., Castro-Urda, V., Fernandez-Lozano, I., Perez, G. J., Scornik, F. S. and Brugada, R.(2013). A novel missense mutation, I890T, in the pore region of cardiac sodium channel causes brugada syndrome. PLoS One 8(1): e53220.
Article Information
Copyright
© 2013 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Tarradas, A., Selga, E., Riuró, H., Scornik, F., Brugada, R. and Vergés, M. (2013). Cell Surface Protein Biotinylation and Analysis. Bio-protocol 3(16): e857. DOI: 10.21769/BioProtoc.857.
Category
Cell Biology > Cell structure > Cell surface
Biochemistry > Protein > Isolation and purification
Biochemistry > Protein > Labeling
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









