- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Determination of Ferric Chelate Reductase Activity in the Arabidopsis thaliana Root
Published: Vol 3, Iss 15, Aug 5, 2013 DOI: 10.21769/BioProtoc.843 Views: 16261
Reviewed by: Tie Liu

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

A Semi-throughput Procedure for Assaying Plant NADP-malate Dehydrogenase Activity Using a Plate Reader
Kevin Baudry and Emmanuelle Issakidis-Bourguet
Aug 20, 2023 1474 Views

An in vitro Assay to Probe the Formation of Biomolecular Condensates
Yu Zhang and Shen Lisha
Sep 5, 2023 3209 Views

Immunofluorescence for Detection of TOR Kinase Activity In Situ in Photosynthetic Organisms
Ana P. Lando [...] Giselle M. A. Martínez-Noël
Dec 20, 2024 1824 Views
Abstract
Plants have developed two distinct mechanisms, i.e., strategy I (reduction strategy) and II (chelation strategy), to mobilize insoluble Fe(III) in the rhizosphere and transport it through the plasma membrane. Arabidopsis thaliana and other dicots rely on strategy I. In this strategy, the rhizosphere is first acidified by a PM-localized H+-ATPase, AHA2. Then, FERRIC CHELATE REDUCTASE 2 (FRO2) reduces Fe(III) to soluble Fe(II). Finally, the reduced Fe is taken up by a high-affinity transporter, IRON-REGULATED TRANSPORTER 1 (IRT1). Root ferric chelate reductase activity can be quantified spectrophotometrically by the formation of Purple-colored Fe(II)-ferrozine complex in darkness.
Keywords: Arabidopsis thalianaMaterials and Reagents
- Arabidopsis thaliana plants [wild-type Col-0 and T-DNA insertion line of FERRIC REDUCTASE DEFECTIVE 3 (frd3-1) are used as examples below]
- Murashige and Skoog (MS) salts
- Ethylenediaminetetraacetic acid ferric sodium salt [Fe(III)-EDTA] (Sigma-Aldrich, catalog number: E6760 )
- 3-(2-Pyridyl)-5,6-diphenyl-1,2,4-triazine-4’,4”-disulfonic acid sodium salt (Ferrozine) (Sigma-Aldrich, catalog number: P9762 )
- Assay solution (see Recipes)
Equipment
- 1.5 ml Eppendorf tubes
- Spectrophotometer (Shimadzu, model: UV-1700 )
Procedure
- Col-0 and frd3-1 seeds were placed on media containing 1/4 Murashige and Skoog (MS) salts, 50 μM Fe-EDTA, 0.5% sucrose, and 1.5% agar (basal medium).
- After stratification for 2 days at 4 °C, the plates were kept in a growth incubator under a long-day photoperiod (16 h light, 8 h darkness) at 25 °C.
- Fe deficiency was applied by transferring 7-day-old seedlings to basal medium without Fe-EDTA but containing 300 μM ferrozine [3-(2-pyridyl)-5,6-diphenyl-1,2,4-triazine sulfonate]. Then, the plants were grown for additional three days on this medium.
- 700 μl of assay solution is placed in a 1.5 ml eppendorf tube, tube is placed onto scale and the weight of the tube is tared (zeroed).
- Both primary and lateral roots of five plants are soaked totally in this assay solution in order to prevent their drying, the tube is weighed again and the fresh weight of the sample is recorded.
Notes:- The assay solution should be kept at dark during the experiment.
- Maximum fresh weight of the roots recommended for this assay is 200 mg.
- Roots are not cut into pieces.
- The assay solution should be kept at dark during the experiment.
- The tube is mixed by tapping several times for increasing the contact of roots with assay solution, and incubated for 30 min in darkness at room temperature.
- At the end of the incubation, purple-colored Fe(II)-ferrozine complex formation is observed around the roots in the solution (Figure 1a).
Note: Much deeper purple color formation is observed around the roots of the plants treated with Fe deficiency (Figure 1b).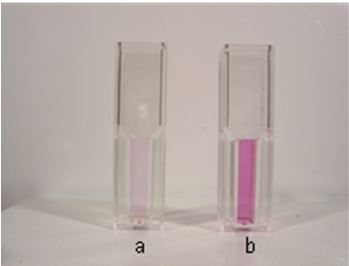
Figure 1. Purple-colored Fe(II)-ferrozine complex formation of assay solution before and after Fe deficiency. - The absorbance of the assay solution is determined in a spectrophotometer at 562 nm against an identical assay solution without any plants (blank).
- Purple-colored Fe(II)-ferrozine complex formation is quantified using a molar extinction coefficient of 28.6 mM-1 cm-1 as in the equation of
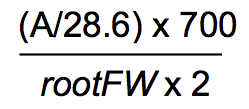
- The experimental results are presented in the unit of μM Fe(II)/g root FW/hr as the mean of three biological repeats with six technical replicates each (Figure 2).
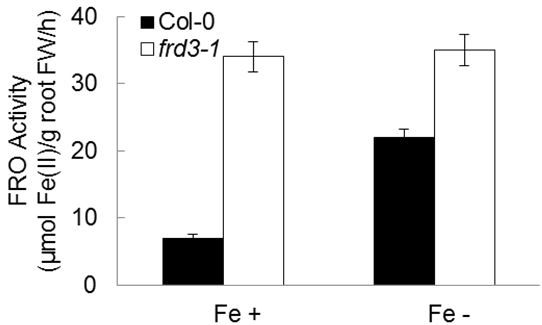
Figure 2. Ferric Chelate Reductase activity in roots of Col-0 and frd3-1 under Fe-sufficient or -deficient conditions.
Recipes
- Assay solution
The assay solution is composed of 0.1 mM Fe(III)-EDTA and 0.3 mM ferrozine in distilled water. Prepare fresh before each experiment and kept at dark.
Acknowledgments
This protocol is adapted from Yi and Guerinot (1996).
References
- Yi, Y. and Guerinot, M. L. (1996). Genetic evidence that induction of root Fe(III) chelate reductase activity is necessary for iron uptake under iron deficiency. Plant J 10(5): 835-844.
Article Information
Copyright
© 2013 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Aksoy, E. and Koiwa, H. (2013). Determination of Ferric Chelate Reductase Activity in the Arabidopsis thaliana Root. Bio-protocol 3(15): e843. DOI: 10.21769/BioProtoc.843.
Category
Plant Science > Plant biochemistry > Protein > Activity
Biochemistry > Protein > Activity
Cell Biology > Cell staining > Iron
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link