- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Analyzing Inhibitory Effects of Reagents on Mycoplasma Gliding and Adhesion
Published: Vol 3, Iss 14, Jul 20, 2013 DOI: 10.21769/BioProtoc.829 Views: 11944
Reviewed by: Fanglian He

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Bacterial Pathogen-mediated Suppression of Host Trafficking to Lysosomes: Fluorescence Microscopy-based DQ-Red BSA Analysis
Mădălina Mocăniță [...] Vanessa M. D'Costa
Mar 5, 2024 2902 Views

Purification of Native Dentilisin Complex from Treponema denticola by Preparative Continuous Polyacrylamide Gel Electrophoresis and Functional Analysis by Gelatin Zymography
Pachiyappan Kamarajan [...] Yvonne L. Kapila
Apr 5, 2024 2095 Views

In Silico Prediction and In Vitro Validation of Bacterial Interactions in the Plant Rhizosphere Using a Synthetic Bacterial Community
Arijit Mukherjee [...] Sanjay Swarup
Nov 5, 2025 1671 Views
Abstract
Dozens of Mycoplasma species bind to solid surfaces and glide in the direction of the membrane protrusion at a pole. In gliding, Mycoplasma legs catch, pull and release sialylated oligosaccharides fixed on a solid surface. The analyses of inhibitory effects of sialylated compounds on gliding of Mycoplasma can determine the target structure of Mycoplasma for gliding and adhesion.
Keywords: Sialylated oligosaccharideMaterials and Reagents
- M. mobile 163K strain
- M. pneumoniae M129 strain
- Horse serum (Life Technologies, catalog number: 16050-122 )
- Interested reagents (for example, 0.05-1 mM 3’-sialyllactose, Dextra Laboratories, catalog number: SL302 )
- Heart infusion broth (Becton, Dickinson and Company, catalog number: 238400 )
- Yeast extract (Becton, Dickinson and Company, catalog number: 212750 )
- Amphotericin B (Sigma-Aldrich, catalog number: A2942 )
- Ampicillin (Nacalai Tesque, catalog number: 02739-32 )
- NaCl
- Sodium phosphate (pH 7.3)
- Glucose
- Aluotto medium (see Recipes)
- PBS-G buffer (see Recipes)
Equipment
- Test tubes
- Tissue culture flask
- Scraper (Greiner Bio-One GmbH, catalog number: 541070 )
- 0.45-μm-pore size filter (Millipore, catalog number: SLHVX13NK ) (PolyVinylidene DiFluoride, or equivalent)
- 25 °C incubator (Yamato, model: IL62 )
- 37 °C incubator (Yamato, model: IS62 )
- Centrifuges (Sigma Zentrifugen, model: SIGMA 1-14 )
- Optical microscope (OLYMPUS, model: BX51 )
- Stage heater (for M. pneumoniae) (Minitube, model: HT200 )
- Lens heater (for M. pneumoniae) (Tokai Hit, model: MATS-LH )
Software
- ImageJ
Procedure
- M. mobile
- Cultivate M. mobile cells in a growth medium (Aluotto medium) at 25 °C incubator to mid log phase.
- Collect the cells in mid log phase at 0.03-0.08 of OD600, by centrifugation at 13,000 x g for 4 min at RT and suspend them in the Aluotto medium to be OD600 = 1.0.
- Wash the cells three times by centrifugation followed by suspension, and resuspend the cells with PBS-G buffer by the same volume with the Aluotto medium used in A-2.
- Prepare a tunnel chamber (5 mm interior width, 18 mm length, 60 μm wall thickness) composed of a coverslip, a glass slide, and double-sided tapes (see Figure 1).
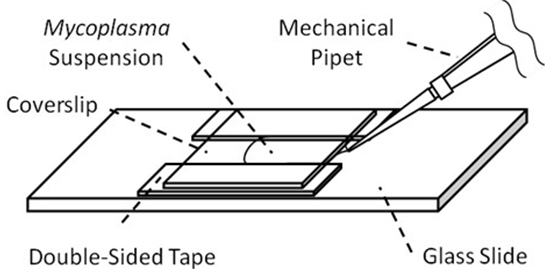
Figure 1. Tunnel chamber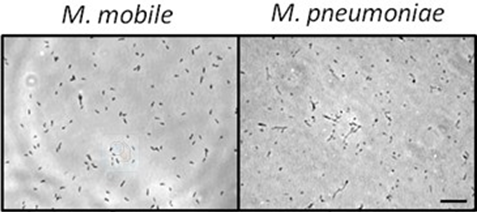
Figure 2. Appearance of Mycoplasma cells (Bar 10 mm) - Insert 10-50 μl of the cell suspension into the tunnel chamber to fill the tunnel.
- Observe the cells bound to the coverslip with microscope with a 100x phase-contrast objective lens with video recording (see Figure 2 left).
- Replace the PBS-G buffer with the buffer containing the interested reagents (i.e. sialylated compounds, monoclonal antibody, etc.). Put the buffer on one side of tunnel and suck the buffer from the other side using a filter paper.
- Count the number of bound cells by a command “analyze > analyze particles” of ImageJ, an image analyzing software. Unbound cells are in Brownian motion and cannot be counted by this Image J command, owing to the insufficient image density.
- Cultivate M. mobile cells in a growth medium (Aluotto medium) at 25 °C incubator to mid log phase.
- M. pneumoniae
- Cultivate M. pneumoniae cells in Aluotto medium at 37 °C incubator to mid log phase. The density of cells is not detectable because the cells are not floating in the medium. However, the density in mid log phase should be 0.02-0.05 at OD600, if the cells are suspended into the medium.
- Replace the medium by 2-5 times smaller volume of PBS containing 10% horse serum.
- Scrape the bottom of culture flask to release Mycoplasma cells into the solution, because the cells adhere to the flask bottom tightly.
- Recover the cell suspension.
- Filter the suspension through a membrane filter unit with a 0.45 μm-pore size.
- Prepare a tunnel chamber.
- Insert 10-50 μl of the cell suspension into the tunnel chamber to fill the inside.
- Incubate the tunnel chamber for 60 min, with facing of coverslip-side to the stage heater. The water evaporates only slightly in Japanese climate, but the tunnel chamber should be cover by a lid of Petri dish.
- Observe the cells bound to the coverslip by the microscope with a 100x phase-contrast objective lens heated by the lens heater, with video recording (see Figure 2 right).
- Replace the buffer with the buffer containing the interested reagents. Put the buffer on one side of tunnel and suck the buffer from the other side using a filter paper.
- Count the number of bound cell by a command “analyze > analyze particles” of ImageJ, an image analyzing software.
- Cultivate M. pneumoniae cells in Aluotto medium at 37 °C incubator to mid log phase. The density of cells is not detectable because the cells are not floating in the medium. However, the density in mid log phase should be 0.02-0.05 at OD600, if the cells are suspended into the medium.
Recipes
- Aluotto medium
2.1% heart infusion broth
0.56% yeast extract
10% horse serum
0.025% amphotericin B
0.005% ampicillin - PBS-G buffer
68 mM NaCl
75 mM sodium phosphate (pH 7.3)
10 mM glucose
Acknowledgments
This protocol is adapted from Kasai et al. (2013).
References
- Kasai, T., Nakane, D., Ishida, H., Ando, H., Kiso, M. and Miyata, M. (2013). Role of binding in Mycoplasma mobile and Mycoplasma pneumoniae gliding analyzed through inhibition by synthesized sialylated compounds. J Bacteriol 195(3): 429-435.
Article Information
Copyright
© 2013 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Kasai, T. and Miyata, M. (2013). Analyzing Inhibitory Effects of Reagents on Mycoplasma Gliding and Adhesion. Bio-protocol 3(14): e829. DOI: 10.21769/BioProtoc.829.
Category
Microbiology > Microbe-host interactions > Bacterium
Cell Biology > Cell structure > Cell adhesion
Cell Biology > Cell movement > Cell motility
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











