- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
EdU labeling of Trypanosome Cells and Their Kinetoplast DNA (kDNA)
Published: Vol 3, Iss 12, Jun 20, 2013 DOI: 10.21769/BioProtoc.799 Views: 10935

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Molecular Surveillance of Malaria Using the PF AmpliSeq Custom Assay for Plasmodium falciparum Parasites from Dried Blood Spot DNA Isolates from Peru
Johanna Helena Kattenberg [...] Anna Rosanas-Urgell
Mar 5, 2023 2150 Views

Rapid Plasmid-Free Generation of Recombinant Positive-Strand RNA Viruses That Use IRES-Mediated Translation Using an Expansion of the Circular Polymerase Extension Reaction (CPER)
Hirotaka Yamamoto [...] Takausuke Fukuhara
Apr 20, 2025 2083 Views

Implementation of Fusion Primer-Driven Racket PCR Protocol for Genome Walking
Yinwei Gu [...] Haixing Li
Dec 5, 2025 941 Views
Abstract
Trypanosome mitochondrial genome, known as Kinetoplast DNA (kDNA), is a massive network of interlocked DNA rings. The studies of kDNA replication and architecture are of major significance since kDNA is a valid drug target. However, DNA in procyclic trypanosomes can not be labeled with tracer concentrations of 3[H]-thymidine, possibly because they lack a high-affinity transporter for thymidine. Therefore, BrdU, a thymidine analog, has been used at high concentrations to study kDNA replication. However, the detection of BrdU with anti-BrdU antibody requires harsh conditions such as the acid or heat treatment to seperate double DNA strand, which affects the ability for other antibodies to bind as well as the morphology and ability for dyes that require dsDNA to bind efficiently. Instead, EdU (5-Ethynyl-2′-deoxyuridine), a novel thymidine analog, can be used to study kDNA replication and cell proliferation with a simplified protocol. Detection of EdU is based on a click reaction, which is a copper (I) catalyzed reaction between an azide and an alkyne. This click reaction does not require DNA denaturation and it is multiplex compatible, such as other antibodies and dyes for cell cycle analyses. To visualize trypanosome replicating nuclear DNA and kDNA, EdU is added into the medium of cell culture and incubated for 0.5 h to 3 h and then detected by the following procedures.
Keywords: TrypanosomeMaterials and Reagents
- Click-iT® Cell Reaction Buffer Kit (Life Technologies, InvitrogenTM, catalog number: C10269 )
- EdU (Life Technologies, InvitrogenTM, catalog number: A10044 )
- Alexa Fluor® 488 azide (Life Technologies, InvitrogenTM, catalog number: A10266 )
OR Alexa Fluor® 594 azide (Life Technologies, InvitrogenTM, catalog number: C10270 )
- 4% Paraformaldehyde (PFA) in PBS
- Deionized water (ddH2O)
- Poly-L-Lysine (Sigma-Aldrich, catalog number: P8920 )
- TEFLON printed slides 8-well, 6 mm diameter (Electron Microscopy Sciences)
- Vectashield® mounting medium with DAPI (Vector Laboratories, catalog number: H-1200 )
- Nail polish
- EdU solution (molecular weight of EdU: 252.22 g/mol) (see Recipes)
- 0.1 M Glycine in 1x PBS (see Recipes)
Equipment
- A humid chamber (a box with moistened paper tower; can protect from light and at least 8 x 10 cm2)
- Microscope Coverslips, 22 x 50 cm2 (Fisherbrand®)
- Centrifuge
- 28 °C 5% CO2 Cell culture incubator
- Fluorescent microscope
Procedure
I. EdU labeling of trypanosome cells
Notes:
1)This protocol has not been used for the bloodstream form of trypanosomes yet. However, it is supposed to work well for the bloodstream form of trypanosomes too.
2)From step 3 to the end of the protocol, operate at room temperature (RT).
3)From step 9, please protect from light by covering the box with lid or the eppendorf tubes with foil paper. In steps 7, 8, and 10, use a 100 or 200 μl pipette to remove solution from each well.
- Culture procyclic trypanosomes in 10 ml SDM-79 medium supplemented with 10% FBS at 28 °C, 5% CO2 incubator.
- Add EdU solution to cell culture at final concentration of 50 μM to 100 μM and continue to incubate at the same condition as step 1 for 30 min to 3 h as planned.
- Trypanosome cells are then pelleted at 700 x g for 5 min.
- Wash the cells with 1 ml 1x PBS once and resuspend cells in 1x PBS at around 2 x 107 cells/ml.
- Coat the 8-well glass slide with poly-L-lysine (0.1% w/v in H2O).
- Add 25 μl cell suspension onto each well in a humid chamber (see Equipment 1) and seat for 5 min.
- Remove cell suspension and add 40 μl 4% paraformaldehyde to fix cells for 5 min.
- Remove fixatives and wash cells twice with 50 μl 0.1 M Glycine for 5 min.
- Prepare Click-iT reaction cocktail (make cocktail fresh and keep dark).
For 200 μl (1 slide or 8 wells, 25 μl/well):
Component A: 1x TBS 176 μl
Component B: 100 mM CuSO4 4 μl
Component C: Buffer additive 20 μl
1 mM Alexa-488-azide 1 μl
(OR 1 mM Alexa-594-azide 1 μl)
- Remove the wash buffer from each well at step 8 and add 25 μl reaction cocktail per well and incubated for 1 h at RT. Protect from light.
- After 1 h incubation, remove the reaction cocktail, wash twice with 50 μl 1x PBS. Protect from light.
- Mount slides with Vectashield Mounting Medium with DAPI (1.5 μg/ml), seal with nail polish and seat for 15~30 min. Protect from light.
- Examine by Fluorescent Microscopy (using 63x or 100x objective lens) (Figure 1).

Figure 1. EdU labeling of procyclic form of trypanosome cells. DNA synthesis was measured by adding 100 μM EdU to the culture medium for 1 h before harvest. Cells were then adhered to a poly-L-lysine coated 8-well slide and EdU was detected with Alexa-488-azide followed by the counterstaining of DNA and nucleus with 1.5 μg/ml DAPI. The inset in the EdU panel is the enlarged EdU labeling image of the cell on the right in the Phase panel. N, nucleus; k, kDNA. Red, DAPI; Green, EdU. Bar, 5 μm.
II. EdU labeling of kDNA networks isolated from trypanosomes
Notes:
1)From step 3 to the end of the protocol, operate at RT (room temperature).
2)From step 7, please protect from light by covering the box with lid or the eppendorf tubes with foil paper.
3) In steps 8 and 9, please use a 100 or 200 μl pipette to remove solution from each well.
- Culture the procyclic trypanosomes in SDM-79 medium supplemented with 10% FBS at 28 °C, 5% CO2 incubator. For kDNA isolation, prepare more than 1 x 108 cells in total.
- Add EdU solution to cell culture at 50 μM to 100 μM and continue to incubate for 30 min to 3 h as planned.
- Isolate kDNA networks from ≥ 5 x 107 trypanosome cells.
- Coat the 8-well glass slide with 1:50 diluted poly-L-lysine in ddH2O.
- Mix 1 μl isolated kDNA networks with 19 μl 1x PBS (add 1x PBS in each well first and then mix kDNA with 1x PBS on the well).
Note: Before doing EdU labeling of isolated kDNA networks, it is suggested to examine kDNA networks abundance by DAPI staining only by following steps 5, 6, 10 and 11 (at least 15 kDNA networks in each field to continue).
- Allow kDNA networks seat on the slide (poly-L-lysine coated) for 30 min at room temperature in humid chamber (see Equipment 1).
- Prepare Click-iT reaction cocktail (make cocktail fresh and keep dark).
For 200 μl (1 slide or 8 wells, 25 μl/well):
Component A: 1x TBS 176 μl
Component B: 100 mM CuSO4 4 μl
Component C: Buffer additive 20 μl
1 mM Alexa-488-azide 1 μl
(OR 1 mM Alexa-594-azide 1 μl)
- Remove kDNA and 1x PBS mixture from each well and add 25 μl reaction cocktail per well and incubated for 1 h at RT. Protect from light.
- After 1 h incubation, remove the reaction cocktail, wash twice with 50 μl 1x PBS. Protect from light.
- Mount slides with Vectashield Mounting Medium with DAPI (1.5 μg/ml), and seal with nail polish, and seat for 15~30 min. Protect from light.
- Examine by Fluorescent Microscopy (using 100x objective lens) (Figure 2).
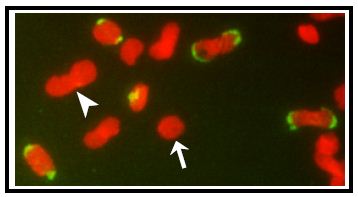
Figure 2. EdU labeling of isolated kDNA network from trypanosome cells. Networks were isolated from EdU-labeled cells and adhered to an 8-well glass slide and incorporated EdU was then detected with Alexa-488-azide (in green) and networks were stained with 1.5 μg/ml DAPI (in red). Arrows, a unit-sized pre-replication kDNA; Arrowhead, a double-sized post-replication kDNA.
Recipes
- EdU solution (molecular weight of EdU: 252.22 g/mol)
Dissolve 12.16 mg EdU in 1 ml ddH2O for 50 mM EdU solution
Dissolve 12.16 mg EdU in 1 ml ddH2O for 50 mM EdU solution
- 1x PBS
Dissolve the following in 800 ml distilled H2O
8 g of NaCl
0.2 g of KCl
1.44 g of Na2HPO4
0.24 g of KH2PO4
Adjust pH to 7.4; then adjust volume to 1 L with additional distilled H2O. Sterilize by autoclaving.
Acknowledgments
I thank all lab members of Paul Englund and Robert Jensen for helpful discussions. This work was supported by NIH grant AI058613.
References
- Wang, J., Englund, P. T. and Jensen, R. E. (2012). TbPIF8, a Trypanosoma brucei protein related to the yeast Pif1 helicase, is essential for cell viability and mitochondrial genome maintenance. Mol Microbiol 83(3): 471-485.
Article Information
Copyright
© 2013 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Wang, J. (2013). EdU labeling of Trypanosome Cells and Their Kinetoplast DNA (kDNA). Bio-protocol 3(12): e799. DOI: 10.21769/BioProtoc.799.
Category
Microbiology > Microbial genetics > DNA > DNA labeling
Molecular Biology > DNA > DNA synthesis
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









