- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
High Resolution Detection of Genetic Changes Associated with Transposons
Published: Vol 3, Iss 11, Jun 5, 2013 DOI: 10.21769/BioProtoc.788 Views: 10412
Reviewed by: Tie Liu

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Experimental Pipeline for SNP and SSR Discovery and Genotyping Analysis of Mango (Mangifera indica L.)
Michal Sharabi-Schwager [...] Ron Ophir
Aug 20, 2016 11559 Views

Investigation of Transposon DNA Methylation and Copy Number Variation in Plants Using Southern Hybridisation
Vivek Hari Sundar G. and P. V. Shivaprasad
Jun 5, 2022 3412 Views

CAPS-Based SNP Genotyping for Nitrogen-Response Phenotypes in Maize Hybrids
Jannis Jacobs [...] Peter K. Lundquist
Dec 20, 2025 550 Views
Abstract
Transposable elements (TEs) are repetitive sequences, capable of inducing genetic mutations through their transpositional activity, or by non-homologous or illegitimate recombination. Because of their similarity and often high copy numbers, examining the effects of mutations caused by TEs in different samples (tissues, individuals, species, etc.) can be difficult. Thus, high throughput methods have been developed for genotyping TEs in un-sequenced genomes. A common method is termed Transposon Display (or transposon SSAP), which utilizes restriction enzymes and PCR amplification to produce chimeric DNA molecules that include genomic and TE DNA. The advent of second generation sequencing technologies, such as 454-pyrosequencing, have dramatically improved the resolution of this assay, allowing the simultaneous sequencing of all PCR products, representing all amplified TE sites in a specific genome.
Keywords: TransposonMaterials and Reagents
- Two oligonucleotides which form the double-stranded adapter, with an overhang complementary to the overhand of the restriction enzyme used. In the case of Mse I, the overhang is a 5' TA, and the adapter sequences are 5'- TACTCAGGACTCAT-3' and 5'- GACGATGAGTCCTGAG-3'. The two nucleotides at the 5' end of the former oligonucleotide will constitute the 5' TA overhang, after hybridization of the two sequences (green rectangles in Figure 1). These oligonucleotides should be designed such that they do not resemble know sequences in the examined species.
- Pre-selective primers, one complementary to the adapter with the addition of a C nucleotide at the 3' end (5'-GATGAGTCCTGAGTAAC-3'; primer P2 in Figure 1), and the other complementary to the TE of interest (primer P1 in Figure 1). The TE-specific primer should be designed as a reverse-complement of the 5' end of the TE with a Tm=60 °C, between 30-50 bp into the TE (to allow for sequence validation in downstream assays). Restriction enzyme recognition sites (TTAA) between the primer and the 5' end of the TE should be avoided. Both the adapter- and TE-specific primers should be designed with the linker A and B sequences (for 454-pyrosequencing) at their 5' ends.
- NaCl
- T4 DNA ligase and buffer (New England Biolabs, catalog number: M0202 )
- Restriction enzyme MseI (New England Biolabs, catalog number: R0525 )
- Taq DNA polymerase and Taq DNA polymerase buffer (EURx, catalog number: E2500 )
- MgCl2
- dNTP mix

Figure 1. An overview of the TD-454 pyrosequencing method. (a) The genomic samples are fragmented with a restriction enzyme and ligated to adapters (green rectangles); (b) The fragments undergo PCR with a primer specific to the adapter (containing a 454 linker sequence) and a primer specific to the analyzed TE (containing a 454 linker sequence); (c) The resulting PCR amplicons are sequenced with a 454 pyrosequencer.
Equipment
- Thermal cycler
Procedure
- Adapter pair preparation
- Mix the two adapter oligonucleotides to a final concentration of 250 ng/μl.
- Incubate them at 95 °C for 5 min and then at room temperature for 10 min.
- Mix the two adapter oligonucleotides to a final concentration of 250 ng/μl.
- Restriction/Ligation
- Add to a 0.2 ml tube: 1 μl of 10x ligase buffer, 1 μl of 0.5 M NaCl, 1 μl of the adapter pair, 120 units of T4 ligase, 2 units of Mse I, 300-500 ng of genomic DNA and ddH2O to a final volume of 10 μl.
- Mix well and incubate at 37 °C for 2-3 h.
- Dilute reaction 1:10 by adding 90 μl of ddH2O.
- This reaction can be stored at -20 °C.
- Add to a 0.2 ml tube: 1 μl of 10x ligase buffer, 1 μl of 0.5 M NaCl, 1 μl of the adapter pair, 120 units of T4 ligase, 2 units of Mse I, 300-500 ng of genomic DNA and ddH2O to a final volume of 10 μl.
- Pre-selective amplification
- Add to a 0.2 ml tube: 2 μl of 10x Taq DNA polymerase buffer, 2 μl of 25 mM MgCl2, 0.8 μl of dNTP mix, 1 unit of Taq DNA polymerase, 1 μl of 50 ng/μl adapter-specific pre-selective primer,1 μl of 50 ng/ul transposon-specific primer, 4 μl of Restriction/Ligation reaction products (cut with Mse I) and ddH2O to a final volume of 20 μl.
- Use the thermal cycler to PCR with the following program:
- 94 °C for 3 min
- 94 °C for 30 sec
- 60 °C for 30 sec
- 72 °C for 1 min
Return to stage b for 29 times.
- 94 °C for 3 min
- Run the relevantly-sized PCR products (usually a narrow band between 150-550 bp, such as 300-500 bp, depending on the expected number of amplicons) in a 454-pyrosequencing machine (Figure 2).
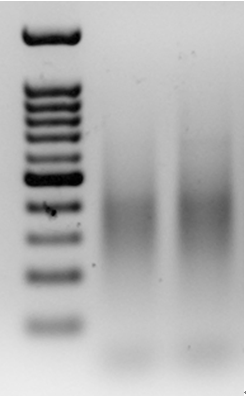
Figure 2. An example of PCR products from pre-selective amplification, using a primer from the adapter and a prom from a Stowaway-like MITE, called Eos, on a 1.5% agarose gel. The lanes include a 100 bp marker (left), pre-selective amplification of goatgrass Aegilops longissima (accession TL05; middle lane) and pre-selective amplification of diploid wheat Triticum urartu (accession TMU06; right lane).
- Add to a 0.2 ml tube: 2 μl of 10x Taq DNA polymerase buffer, 2 μl of 25 mM MgCl2, 0.8 μl of dNTP mix, 1 unit of Taq DNA polymerase, 1 μl of 50 ng/μl adapter-specific pre-selective primer,1 μl of 50 ng/ul transposon-specific primer, 4 μl of Restriction/Ligation reaction products (cut with Mse I) and ddH2O to a final volume of 20 μl.
Acknowledgments
The transposon (TD) display method was adapted from the amplified fragment length polymorphism (AFLP) (Vos et al., 1995 Nucl Acid Res 23:4407-4414). This work was supported by a grant from the Israel Science Foundation (grant # 142/08) to Khalil Kashkush.
References
- Yaakov, B. and Kashkush, K. (2012). Mobilization of Stowaway-like MITEs in newly formed allohexaploid wheat species. Plant Mol Biol 80(4-5): 419-427.
Article Information
Copyright
© 2013 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Yaakov, B. and Kashkush, K. (2013). High Resolution Detection of Genetic Changes Associated with Transposons. Bio-protocol 3(11): e788. DOI: 10.21769/BioProtoc.788.
Category
Systems Biology > Genomics > Transposons
Plant Science > Plant molecular biology > DNA > Genotyping
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










