Improve Research Reproducibility A Bio-protocol resource
- Protocols
- Articles and Issues
- About
- Become a Reviewer
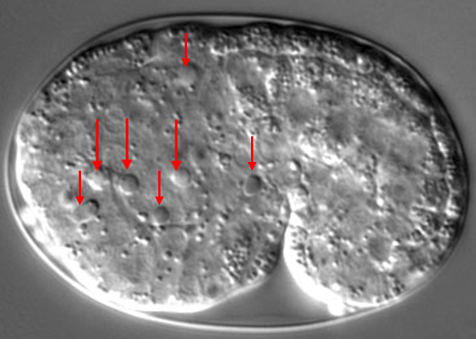
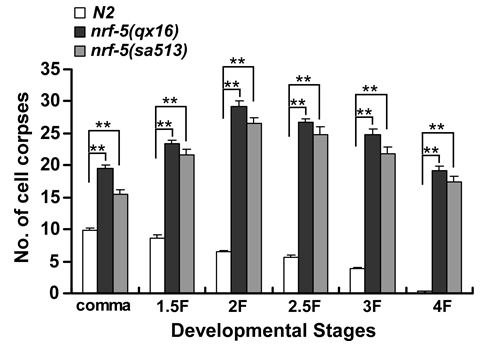
Quantification of Cell Corpses, Cell Death Occurrence, Cell Corpse Duration
Published: Vol 3, Iss 9, May 5, 2013 DOI: 10.21769/BioProtoc.693 Views: 11740
How to cite
Favorite
Cited by